Mechanisms of base selection by human single-stranded selective monofunctional uracil-DNA glycosylase
- PMID: 19324873
- PMCID: PMC2708880
- DOI: 10.1074/jbc.M807846200
Mechanisms of base selection by human single-stranded selective monofunctional uracil-DNA glycosylase
Abstract
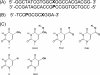
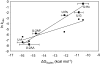
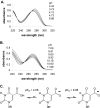
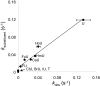
hSMUG1 (human single-stranded selective monofunctional uracil-DNA glyscosylase) is one of three glycosylases encoded within a small region of human chromosome 12. Those three glycosylases, UNG (uracil-DNA glycosylase), TDG (thymine-DNA glyscosylase), and hSMUG1, have in common the capacity to remove uracil from DNA. However, these glycosylases also repair other lesions and have distinct substrate preferences, indicating that they have potentially redundant but not overlapping physiological roles. The mechanisms by which these glycosylases locate and selectively remove target lesions are not well understood. In addition to uracil, hSMUG1 has been shown to remove some oxidized pyrimidines, suggesting a role in the repair of DNA oxidation damage. In this paper, we describe experiments in which a series of oligonucleotides containing purine and pyrimidine analogs have been used to probe mechanisms by which hSMUG1 distinguishes potential substrates. Our results indicate that the preference of hSMUG1 for mispaired uracil over uracil paired with adenine is best explained by the reduced stability of a duplex containing a mispair, consistent with previous reports with Escherichia coli mispaired uracil-DNA glycosylase. We have also extended the substrate range of hSMUG1 to include 5-carboxyuracil, the last in the series of damage products from thymine methyl group oxidation. The properties used by hSMUG1 to select damaged pyrimidines include the size and free energy of solvation of the 5-substituent but not electronic inductive properties. The observed distinct mechanisms of base selection demonstrated for members of the uracil glycosylase family help explain how considerable diversity in chemical lesion repair can be achieved.
Figures



References
-
- Sousa M. M., Krokan H. E., Slupphaug G. ( 2007) Mol. Aspects Med. 28, 276– 306 - PubMed
-
- Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N. M., Haug T., Levine D. W., Krokan H. E. ( 1995) Biochemistry 34, 128– 138 - PubMed
-
- Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J. J., Weibauer K., Jiricny J. ( 1996) J. Biol. Chem. 217, 12767– 12774 - PubMed
-
- Pearl L. H. ( 2000) Mutat. Res. 460, 165– 181 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

