Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF
- PMID: 19324881
- PMCID: PMC2682891
- DOI: 10.1074/jbc.M809221200
Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF
Abstract
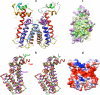
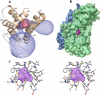
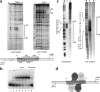
Stenotrophomonas maltophilia is an opportunistic pathogen characterized for its intrinsic low susceptibility to several antibiotics. Part of this low susceptibility relies on the expression of chromosomally encoded multidrug efflux pumps, with SmeDEF being the most relevant antibiotic resistance efflux pump so far studied in this bacterial species. Expression of smeDEF is down-regulated by the SmeT repressor, encoded upstream smeDEF, in its complementary DNA strand. In the present article we present the crystal structure of SmeT and analyze its interactions with its cognate operator. Like other members of the TetR family of transcriptional repressors, SmeT behaves as a dimer and presents some common structural features with other TetR proteins like TtgR, QacR, and TetR. Differing from other TetR proteins for which the structure is available, SmeT turned out to have two extensions at the N and C termini that might be relevant for its function. Besides, SmeT presents the smallest binding pocket so far described in the TetR family of transcriptional repressors, which may correlate with a specific type and range of effectors. In vitro studies revealed that SmeT binds to a 28-bp pseudopalindromic region, forming two complexes. This operator region was found to overlap the promoters of smeT and smeDEF. This finding is consistent with a role for SmeT simultaneously down-regulating smeT and smeDEF transcription, likely by steric hindrance on RNA polymerase binding to DNA.
Figures
Similar articles
-
Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF.Antimicrob Agents Chemother. 2002 Nov;46(11):3386-93. doi: 10.1128/AAC.46.11.3386-3393.2002. Antimicrob Agents Chemother. 2002. PMID: 12384340 Free PMC article.
-
The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia.PLoS Pathog. 2011 Jun;7(6):e1002103. doi: 10.1371/journal.ppat.1002103. Epub 2011 Jun 30. PLoS Pathog. 2011. PMID: 21738470 Free PMC article.
-
A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots.Appl Environ Microbiol. 2014 Aug;80(15):4559-65. doi: 10.1128/AEM.01058-14. Appl Environ Microbiol. 2014. PMID: 24837376 Free PMC article.
-
The TetR family of transcriptional repressors.Microbiol Mol Biol Rev. 2005 Jun;69(2):326-56. doi: 10.1128/MMBR.69.2.326-356.2005. Microbiol Mol Biol Rev. 2005. PMID: 15944459 Free PMC article. Review.
-
The underling mechanism of bacterial TetR/AcrR family transcriptional repressors.Cell Signal. 2013 Jul;25(7):1608-13. doi: 10.1016/j.cellsig.2013.04.003. Epub 2013 Apr 16. Cell Signal. 2013. PMID: 23602932 Review.
Cited by
-
Whole-genome sequence of Stenotrophomonas maltophilia D457, a clinical isolate and a model strain.J Bacteriol. 2012 Jul;194(13):3563-4. doi: 10.1128/JB.00602-12. J Bacteriol. 2012. PMID: 22689246 Free PMC article.
-
Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants.Microorganisms. 2016 Feb 16;4(1):14. doi: 10.3390/microorganisms4010014. Microorganisms. 2016. PMID: 27681908 Free PMC article. Review.
-
The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria.Clin Microbiol Rev. 2015 Apr;28(2):337-418. doi: 10.1128/CMR.00117-14. Clin Microbiol Rev. 2015. PMID: 25788514 Free PMC article. Review.
-
Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa.Antimicrob Agents Chemother. 2014 Jul;58(7):3904-13. doi: 10.1128/AAC.00121-14. Epub 2014 Apr 28. Antimicrob Agents Chemother. 2014. PMID: 24777101 Free PMC article.
-
Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options.Front Microbiol. 2015 Sep 2;6:893. doi: 10.3389/fmicb.2015.00893. eCollection 2015. Front Microbiol. 2015. PMID: 26388847 Free PMC article. Review.
References
-
- WHO (2000) in World Health Organization Report in Infectious Diseases, Geneva
-
- Martinez, J. L., Fajardo, A., Garmendia, L., Hernandez, A., Linares, J. F., Martinez-Solano, L., and Sanchez, M. B. (2009) FEMS Microbiol. Rev. 33 44-65 - PubMed
-
- Vila, J., and Martinez, J. L. (2008) Curr. Drug Targets 9 797-807 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

