Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching
- PMID: 19324899
- PMCID: PMC2685756
- DOI: 10.1093/hmg/ddp151
Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching
Abstract
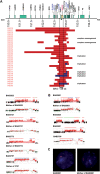
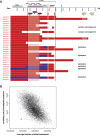
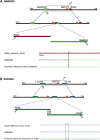
Duplication at the Xq28 band including the MECP2 gene is one of the most common genomic rearrangements identified in neurodevelopmentally delayed males. Such duplications are non-recurrent and can be generated by a non-homologous end joining (NHEJ) mechanism. We investigated the potential mechanisms for MECP2 duplication and examined whether genomic architectural features may play a role in their origin using a custom designed 4-Mb tiling-path oligonucleotide array CGH assay. Each of the 30 patients analyzed showed a unique duplication varying in size from approximately 250 kb to approximately 2.6 Mb. Interestingly, in 77% of these non-recurrent duplications, the distal breakpoints grouped within a 215 kb genomic interval, located 47 kb telomeric to the MECP2 gene. The genomic architecture of this region contains both direct and inverted low-copy repeat (LCR) sequences; this same region undergoes polymorphic structural variation in the general population. Array CGH revealed complex rearrangements in eight patients; in six patients the duplication contained an embedded triplicated segment, and in the other two, stretches of non-duplicated sequences occurred within the duplicated region. Breakpoint junction sequencing was achieved in four duplications and identified an inversion in one patient, demonstrating further complexity. We propose that the presence of LCRs in the vicinity of the MECP2 gene may generate an unstable DNA structure that can induce DNA strand lesions, such as a collapsed fork, and facilitate a Fork Stalling and Template Switching event producing the complex rearrangements involving MECP2.
Figures


References
-
- Van Esch H., Bauters M., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L.R., Gecz J., et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005;77:442–453. - PMC - PubMed
-
- del Gaudio D., Fang P., Scaglia F., Ward P.A., Craigen W.J., Glaze D.G., Neul J.L., Patel A., Lee J.A., Irons M., et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet. Med. 2006;8:784–792. - PubMed
-
- Friez M.J., Jones J.R., Clarkson K., Lubs H., Abuelo D., Bier J.A., Pai S., Simensen R., Williams C., Giampietro P.F., et al. Recurrent infections, hypotonia, and mental retardation caused by duplication of MECP2 and adjacent region in Xq28. Pediatrics. 2006;118:e1687–e1695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

