Distinct functions of elongation factor G in ribosome recycling and translocation
- PMID: 19324963
- PMCID: PMC2673078
- DOI: 10.1261/rna.1592509
Distinct functions of elongation factor G in ribosome recycling and translocation
Abstract
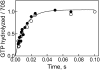
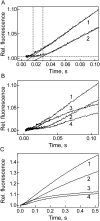
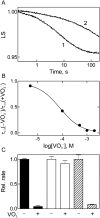
Elongation factor G (EF-G) promotes the translocation step in bacterial protein synthesis and, together with ribosome recycling factor (RRF), the disassembly of the post-termination ribosome. Unlike translocation, ribosome disassembly strictly requires GTP hydrolysis by EF-G. Here we report that ribosome disassembly is strongly inhibited by vanadate, an analog of inorganic phosphate (Pi), indicating that Pi release is required for ribosome disassembly. In contrast, the function of EF-G in single-round translocation is not affected by vanadate, while the turnover reaction is strongly inhibited. We also show that the antibiotic fusidic acid blocks ribosome disassembly by EF-G/RRF at a 1000-fold lower concentration than required for the inhibition of EF-G turnover in vitro and close to the effective inhibitory concentration in vivo, suggesting that the antimicrobial activity of fusidic acid is primarily due to the direct inhibition of ribosome recycling. Our results indicate that conformational coupling between EF-G and the ribosome is principally different in translocation and ribosome disassembly. Pi release is not required for the mechanochemical function of EF-G in translocation, whereas the interactions between RRF and EF-G introduce tight coupling between the conformational change of EF-G induced by Pi release and ribosome disassembly.
Figures


Similar articles
-
Release of ribosome-bound ribosome recycling factor by elongation factor G.J Biol Chem. 2003 Nov 28;278(48):48041-50. doi: 10.1074/jbc.M304834200. Epub 2003 Sep 5. J Biol Chem. 2003. PMID: 12960150
-
Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins--insight into the mechanism of the ribosome recycling step.RNA. 2005 Mar;11(3):275-84. doi: 10.1261/rna.7201805. Epub 2005 Jan 20. RNA. 2005. PMID: 15661844 Free PMC article.
-
Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.J Mol Biol. 2007 Dec 14;374(5):1345-58. doi: 10.1016/j.jmb.2007.10.021. Epub 2007 Oct 16. J Mol Biol. 2007. PMID: 17996252 Free PMC article.
-
Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors.Biochimie. 1996;78(11-12):959-69. doi: 10.1016/s0300-9084(97)86718-1. Biochimie. 1996. PMID: 9150873 Review.
-
Converting GTP hydrolysis into motion: versatile translational elongation factor G.Biol Chem. 2019 Dec 18;401(1):131-142. doi: 10.1515/hsz-2019-0313. Biol Chem. 2019. PMID: 31600135 Review.
Cited by
-
Exit from dormancy in microbial organisms.Nat Rev Microbiol. 2010 Dec;8(12):890-6. doi: 10.1038/nrmicro2453. Epub 2010 Oct 25. Nat Rev Microbiol. 2010. PMID: 20972452 Review.
-
Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome.Genes Dev. 2011 May 1;25(9):917-29. doi: 10.1101/gad.623011. Genes Dev. 2011. PMID: 21536732 Free PMC article.
-
mRNA translocation occurs during the second step of ribosomal intersubunit rotation.Nat Struct Mol Biol. 2011 Apr;18(4):457-62. doi: 10.1038/nsmb.2011. Epub 2011 Mar 13. Nat Struct Mol Biol. 2011. PMID: 21399643 Free PMC article.
-
Key Intermediates in Ribosome Recycling Visualized by Time-Resolved Cryoelectron Microscopy.Structure. 2016 Dec 6;24(12):2092-2101. doi: 10.1016/j.str.2016.09.014. Epub 2016 Nov 3. Structure. 2016. PMID: 27818103 Free PMC article.
-
Similarity and diversity of translational GTPase factors EF-G, EF4, and BipA: From structure to function.RNA Biol. 2016 Dec;13(12):1258-1273. doi: 10.1080/15476286.2016.1201627. Epub 2016 Jun 20. RNA Biol. 2016. PMID: 27325008 Free PMC article. Review.
References
-
- Ali I.K., Lancaster L., Feinberg J., Joseph S., Noller H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
