Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event
- PMID: 19325131
- PMCID: PMC2667025
- DOI: 10.1073/pnas.0900906106
Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event
Abstract
Most flowering plants have been shown to be ancient polyploids that have undergone one or more whole genome duplications early in their evolution. Furthermore, many different plant lineages seem to have experienced an additional, more recent genome duplication. Starting from paralogous genes lying in duplicated segments or identified in large expressed sequence tag collections, we dated these youngest duplication events through penalized likelihood phylogenetic tree inference. We show that a majority of these independent genome duplications are clustered in time and seem to coincide with the Cretaceous-Tertiary (KT) boundary. The KT extinction event is the most recent mass extinction caused by one or more catastrophic events such as a massive asteroid impact and/or increased volcanic activity. These events are believed to have generated global wildfires and dust clouds that cut off sunlight during long periods of time resulting in the extinction of approximately 60% of plant species, as well as a majority of animals, including dinosaurs. Recent studies suggest that polyploid species can have a higher adaptability and increased tolerance to different environmental conditions. We propose that polyploidization may have contributed to the survival and propagation of several plant lineages during or following the KT extinction event. Due to advantages such as altered gene expression leading to hybrid vigor and an increased set of genes and alleles available for selection, polyploid plants might have been better able to adapt to the drastically changed environment 65 million years ago.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
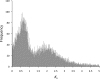
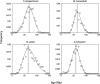
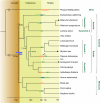
Comment in
-
Surviving the K-T mass extinction: new perspectives of polyploidization in angiosperms.Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5455-6. doi: 10.1073/pnas.0901994106. Epub 2009 Mar 31. Proc Natl Acad Sci U S A. 2009. PMID: 19336584 Free PMC article. No abstract available.
-
Picking up the Ball at the K/Pg Boundary: The Distribution of Ancient Polyploidies in the Plant Phylogenetic Tree as a Spandrel of Asexuality with Occasional Sex.Plant Cell. 2017 Feb;29(2):202-206. doi: 10.1105/tpc.16.00836. Epub 2017 Feb 17. Plant Cell. 2017. PMID: 28213362 Free PMC article. No abstract available.
References
-
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. - PubMed
-
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. - PubMed
-
- Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

