Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum
- PMID: 19325468
- PMCID: PMC2747356
- DOI: 10.1097/CCM.0b013e31819fcc08
Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum
Abstract
Objectives: To advance our biological understanding of pediatric septic shock, we measured the genome-level expression profiles of critically ill children representing the systemic inflammatory response syndrome (SIRS), sepsis, and septic shock spectrum.
Design: Prospective observational study involving microarray-based bioinformatics.
Setting: Multiple pediatric intensive care units in the United States.
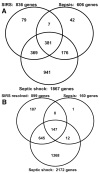
Patients: Children <or=10 years of age: 18 normal controls, 22 meeting criteria for SIRS, 32 meeting criteria for sepsis, and 67 meeting criteria for septic shock on day 1. The available day 3 samples included 20 patients still meeting sepsis criteria, 39 patients still meeting septic shock criteria, and 24 patients meeting the exclusive day 3 category, SIRS resolved.
Interventions: None other than standard care.
Measurements and main results: Longitudinal analyses were focused on gene expression relative to control samples and patients having paired day 1 and day 3 samples. The longitudinal analysis focused on up-regulated genes revealed common patterns of up-regulated gene expression, primarily corresponding to inflammation and innate immunity, across all patient groups on day 1. These patterns of up-regulated gene expression persisted on day 3 in patients with septic shock, but not to the same degree in the other patient classes. The longitudinal analysis focused on down-regulated genes demonstrated gene repression corresponding to adaptive immunity-specific signaling pathways and was most prominent in patients with septic shock on days 1 and 3. Gene network analyses based on direct comparisons across the SIRS, sepsis, and septic shock spectrum, and all available patients in the database, demonstrated unique repression of gene networks in patients with septic shock corresponding to major histocompatibility complex antigen presentation. Finally, analyses focused on repression of genes corresponding to zinc-related biology demonstrated that this pattern of gene repression is unique to patients with septic shock.
Conclusions: Although some common patterns of gene expression exist across the pediatric SIRS, sepsis, and septic shock spectrum, septic shock is particularly characterized by repression of genes corresponding to adaptive immunity and zinc-related biology.
Conflict of interest statement
The authors have not disclosed any potential conflicts of interest.
Figures
Comment in
-
Profiling pediatric sepsis.Crit Care Med. 2009 May;37(5):1795-6. doi: 10.1097/CCM.0b013e3181a1a220. Crit Care Med. 2009. PMID: 19373046 No abstract available.
References
-
- Proulx F, Fayon M, Farrell CA, et al. Epidemiology of sepsis and multiple organ dysfunction syndrome in children. Chest. 1996;109:1033–1037. - PubMed
-
- Shanley TP, Hallstrom C, Wong HR. Sepsis. In: Fuhrman BP, Zimmerman JJ, editors. Pediatric Critical Care Medicine. 3. St. Louis: Mosby; 2006. pp. 1474–1493.
-
- Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6:2–8. - PubMed
-
- Baue AE. MOF, MODS, and SIRS: What is in a name or an acronym? Shock. 2006;26:438 – 449. - PubMed
-
- Baue AE. A debate on the subject “Are SIRS and MODS important entities in the clinical evaluation of patients?” The con position. Shock. 2000;14:590–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

