Accurate prediction of peptide binding sites on protein surfaces
- PMID: 19325869
- PMCID: PMC2653190
- DOI: 10.1371/journal.pcbi.1000335
Accurate prediction of peptide binding sites on protein surfaces
Abstract
Many important protein-protein interactions are mediated by the binding of a short peptide stretch in one protein to a large globular segment in another. Recent efforts have provided hundreds of examples of new peptides binding to proteins for which a three-dimensional structure is available (either known experimentally or readily modeled) but where no structure of the protein-peptide complex is known. To address this gap, we present an approach that can accurately predict peptide binding sites on protein surfaces. For peptides known to bind a particular protein, the method predicts binding sites with great accuracy, and the specificity of the approach means that it can also be used to predict whether or not a putative or predicted peptide partner will bind. We used known protein-peptide complexes to derive preferences, in the form of spatial position specific scoring matrices, which describe the binding-site environment in globular proteins for each type of amino acid in bound peptides. We then scan the surface of a putative binding protein for sites for each of the amino acids present in a peptide partner and search for combinations of high-scoring amino acid sites that satisfy constraints deduced from the peptide sequence. The method performed well in a benchmark and largely agreed with experimental data mapping binding sites for several recently discovered interactions mediated by peptides, including RG-rich proteins with SMN domains, Epstein-Barr virus LMP1 with TRADD domains, DBC1 with Sir2, and the Ago hook with Argonaute PIWI domain. The method, and associated statistics, is an excellent tool for predicting and studying binding sites for newly discovered peptides mediating critical events in biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

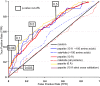

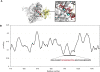
Similar articles
-
Predicting protein-peptide interactions via a network-based motif sampler.Bioinformatics. 2004 Aug 4;20 Suppl 1:i274-82. doi: 10.1093/bioinformatics/bth922. Bioinformatics. 2004. PMID: 15262809
-
MFPred: Rapid and accurate prediction of protein-peptide recognition multispecificity using self-consistent mean field theory.PLoS Comput Biol. 2017 Jun 26;13(6):e1005614. doi: 10.1371/journal.pcbi.1005614. eCollection 2017 Jun. PLoS Comput Biol. 2017. PMID: 28650961 Free PMC article.
-
Analyzing molecular interactions.Curr Protoc Bioinformatics. 2003 May;Chapter 8:Unit8.1. doi: 10.1002/0471250953.bi0801s01. Curr Protoc Bioinformatics. 2003. PMID: 18428708 Review.
-
An integrative approach for predicting interactions of protein regions.Bioinformatics. 2008 Aug 15;24(16):i35-41. doi: 10.1093/bioinformatics/btn290. Bioinformatics. 2008. PMID: 18689837
-
Protein complexes: structure prediction challenges for the 21st century.Curr Opin Struct Biol. 2005 Feb;15(1):15-22. doi: 10.1016/j.sbi.2005.01.012. Curr Opin Struct Biol. 2005. PMID: 15718128 Review.
Cited by
-
Leveraging Machine Learning Models for Peptide-Protein Interaction Prediction.ArXiv [Preprint]. 2024 Feb 7:arXiv:2310.18249v2. ArXiv. 2024. Update in: RSC Chem Biol. 2024 Mar 13;5(5):401-417. doi: 10.1039/d3cb00208j. PMID: 37961736 Free PMC article. Updated. Preprint.
-
Methods for Molecular Modelling of Protein Complexes.Methods Mol Biol. 2021;2305:53-80. doi: 10.1007/978-1-0716-1406-8_3. Methods Mol Biol. 2021. PMID: 33950384 Review.
-
Modelling binding between CCR5 and CXCR4 receptors and their ligands suggests the surface electrostatic potential of the co-receptor to be a key player in the HIV-1 tropism.Retrovirology. 2013 Nov 11;10:130. doi: 10.1186/1742-4690-10-130. Retrovirology. 2013. PMID: 24215935 Free PMC article.
-
Modular binder technology by NGS-aided, high-resolution selection in yeast of designed armadillo modules.Proc Natl Acad Sci U S A. 2024 Jul 2;121(27):e2318198121. doi: 10.1073/pnas.2318198121. Epub 2024 Jun 25. Proc Natl Acad Sci U S A. 2024. PMID: 38917007 Free PMC article.
-
The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold.Open Biol. 2014 Jan 15;4(1):130090. doi: 10.1098/rsob.130090. Open Biol. 2014. PMID: 24430336 Free PMC article.
References
-
- Diella F, Haslam N, Chica C, Budd A, Michael S, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–6603. - PubMed
-
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. - PubMed
-
- Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr Opin Biotechnol. 2006;17:465–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

