Panorama phylogenetic diversity and distribution of Type A influenza virus
- PMID: 19325912
- PMCID: PMC2658884
- DOI: 10.1371/journal.pone.0005022
Panorama phylogenetic diversity and distribution of Type A influenza virus
Abstract
Background: Type A influenza virus is one of important pathogens of various animals, including humans, pigs, horses, marine mammals and birds. Currently, the viral type has been classified into 16 hemagglutinin and 9 neuraminidase subtypes, but the phylogenetic diversity and distribution within the viral type largely remain unclear from the whole view.
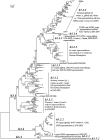
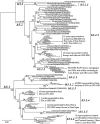
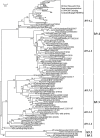
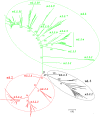
Methodology/principal findings: The panorama phylogenetic trees of influenza A viruses were calculated with representative sequences selected from approximately 23,000 candidates available in GenBank using web servers in NCBI and the software MEGA 4.0. Lineages and sublineages were classified according to genetic distances, topology of the phylogenetic trees and distributions of the viruses in hosts, regions and time.
Conclusions/significance: Here, two panorama phylogenetic trees of type A influenza virus covering all the 16 hemagglutinin subtypes and 9 neuraminidase subtypes, respectively, were generated. The trees provided us whole views and some novel information to recognize influenza A viruses including that some subtypes of avian influenza viruses are more complicated than Eurasian and North American lineages as we thought in the past. They also provide us a framework to generalize the history and explore the future of the viral circulation and evolution in different kinds of hosts. In addition, a simple and comprehensive nomenclature system for the dozens of lineages and sublineages identified within the viral type was proposed, which if universally accepted, will facilitate communications on the viral evolution, ecology and epidemiology.
Conflict of interest statement
Figures







References
-
- Campitelli L, Donatelli I, Foni E, Castrucci MR, Fabiani C, et al. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. - PubMed
-
- Brown IH, Ludwig S, Olsen CW, Hannoun C, Scholtissek C, et al. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J Gen Virol. 1997;78:553–562. - PubMed
-
- Marozin S, Gregory V, Cameron K, Bennett M, Valette M, et al. Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J Gen Virol. 2002;83:735–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

