Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants
- PMID: 19327164
- PMCID: PMC2671507
- DOI: 10.1186/1471-2229-9-37
Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants
Abstract
Background: Although histone deacetylases from model organisms have been previously identified, there is no clear basis for the classification of histone deacetylases under the RPD3/HDA1 superfamily, particularly on plants. Thus, this study aims to reconstruct a phylogenetic tree to determine evolutionary relationships between RPD3/HDA1 histone deacetylases from six different plants representing dicots with Arabidopsis thaliana, Populus trichocarpa, and Pinus taeda, monocots with Oryza sativa and Zea mays, and the lower plants with Physcomitrella patens.
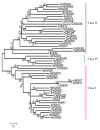
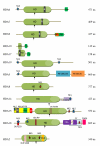
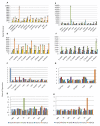
Results: Sixty two histone deacetylases of RPD3/HDA1 family from the six plant species were phylogenetically analyzed to determine corresponding orthologues. Three clusters were formed separating Class I, Class II, and Class IV. We have confirmed lower and higher plant orthologues for AtHDA8 and AtHDA14, classifying both genes as Class II histone deacetylases in addition to AtHDA5, AtHDA15, and AtHDA18. Since Class II histone deacetylases in other eukaryotes have been known to undergo nucleocytoplasmic transport, it remains unknown whether such functional regulation also happens in plants. Thus, bioinformatics studies using different programs and databases were conducted to predict their corresponding localization sites, nuclear export signal, nuclear localization signal, as well as expression patterns. We also found new conserved domains in most of the RPD3/HDA1 histone deacetylases which were similarly conserved in its corresponding orthologues. Assessing gene expression patterns using Genevestigator, it appears that RPD3/HDA1 histone deacetylases are expressed all throughout the plant parts and developmental stages of the plant.
Conclusion: The RPD3/HDA1 histone deacetylase family in plants is divided into three distinct groups namely, Class I, Class II, and Class IV suggesting functional diversification. Class II comprises not only AtHDA5, AtHDA15, and AtHDA18 but also includes AtHDA8 and AtHDA14. New conserved domains have also been identified in most of the RPD3/HDA1 family indicating further versatile roles other than histone deacetylation.
Figures



References
-
- Pandey R, Muller A, Napoli C, Selinger D, Pikaard C, Richards E, Bender J, Mount D, Jorgensen R. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acid Research. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

