Glycoproteomics: past, present and future
- PMID: 19328791
- PMCID: PMC2753369
- DOI: 10.1016/j.febslet.2009.03.049
Glycoproteomics: past, present and future
Abstract
This invited paper reviews the study of protein glycosylation, commonly known as glycoproteomics, beginning with the origins of the subject area in the early 1970s shortly after mass spectrometry was first applied to protein sequencing. We go on to describe current analytical approaches to glycoproteomic analyses, with exemplar projects presented in the form of the complex story of human glycodelin and the characterisation of blood group H eptitopes on the O-glycans of gp273 from Unio elongatulus. Finally, we present an update on the latest progress in the field of automated and semi-automated interpretation and annotation of these data in the form of GlycoWorkBench, a powerful informatics tool that provides valuable assistance in unravelling the complexities of glycoproteomic studies.
Figures

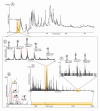
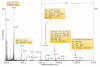

 symbol. Keys related to fragmentation are the following:
symbol. Keys related to fragmentation are the following:  represents a fragmentation on the reducing end side of the glycosidic bond (also known as B or Z ions) and
represents a fragmentation on the reducing end side of the glycosidic bond (also known as B or Z ions) and  represents a fragmentation on the non-reducing end side of the glycosidic bond (also known as C or Y ions).
represents a fragmentation on the non-reducing end side of the glycosidic bond (also known as C or Y ions).References
-
- Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–6. - PubMed
-
- Morris HR, Chalabi S, Panico M, Sutton-Smith M, Clark GF, Goldberg D, Dell A. Glycoproteomics: Past, present and future. International Journal of Mass Spectrometry. 2007;259:16–31.
-
- Morris HR, Thompson MR, Osuga DT, Ahmed AI, Chan SM, Vandenheede JR, Feeney RE. Antifreeze glycoproteins from the blood of an antarctic fish. The structure of the proline-containing glycopeptides. J Biol Chem. 1978;253:5155–62. - PubMed
-
- Taylor GW, Morris HR, Peterson TE, Magnusson S. Advances in Mass Spectrometry. 1980;18:1090.
-
- Morris HR, Panico M, Taylor GW. FAB-mapping of recombinant-DNA protein products. Biochem Biophys Res Commun. 1983;117:299–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

