Domain swapping reveals that low density lipoprotein (LDL) type A repeat order affects ligand binding to the LDL receptor
- PMID: 19329437
- PMCID: PMC2679439
- DOI: 10.1074/jbc.M900194200
Domain swapping reveals that low density lipoprotein (LDL) type A repeat order affects ligand binding to the LDL receptor
Abstract
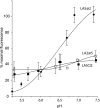
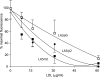
The low density lipoprotein receptor (LDLR) plays a key role in plasma cholesterol homeostasis by binding and internalizing lipoprotein ligands. Studies have revealed that one or more of the seven LDL type A repeats (LA1-LA7) in the receptor are responsible for apolipoprotein binding. In the present study, protein engineering was performed to swap or replace key LA repeats in a recombinant soluble LDLR (sLDLR). Although wild type sLDLR showed strong ligand binding activity, an sLDLR variant in which LA repeat 5 was replaced by a second copy of LA repeat 2 showed low binding activity. Likewise, a variant wherein LA repeats 2 and 5 were swapped displayed low binding activity. At the same time, substitution of LA repeat 2 with a second a copy of repeat 5 resulted in a receptor with ligand binding activity similar to wild type LDLR. When binding assays were conducted with human low density lipoprotein as ligand, LA repeat order was a less important determinant of binding activity. Taken together, the data indicate that the sequential order of LA repeats plays a key role in ligand binding properties of LDLR.
Figures




Similar articles
-
A two-module region of the low-density lipoprotein receptor sufficient for formation of complexes with apolipoprotein E ligands.Biochemistry. 2004 Feb 3;43(4):1037-44. doi: 10.1021/bi035529y. Biochemistry. 2004. PMID: 14744149
-
Conversion of a human low-density lipoprotein receptor ligandbinding repeat to a virus receptor: identification of residues important for ligand specificity.Proc Natl Acad Sci U S A. 1998 Jul 21;95(15):8467-72. doi: 10.1073/pnas.95.15.8467. Proc Natl Acad Sci U S A. 1998. PMID: 9671701 Free PMC article.
-
Molecular studies of pH-dependent ligand interactions with the low-density lipoprotein receptor.Biochemistry. 2008 Nov 4;47(44):11647-52. doi: 10.1021/bi801117t. Epub 2008 Oct 11. Biochemistry. 2008. PMID: 18847225 Free PMC article.
-
Structural features of the low-density lipoprotein receptor facilitating ligand binding and release.Biochem Soc Trans. 2004 Nov;32(Pt 5):721-3. doi: 10.1042/BST0320721. Biochem Soc Trans. 2004. PMID: 15493997 Review.
-
Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis.J Am Coll Cardiol. 2013 Oct 15;62(16):1401-8. doi: 10.1016/j.jacc.2013.07.056. Epub 2013 Aug 21. J Am Coll Cardiol. 2013. PMID: 23973703 Review.
Cited by
-
Interaction between SCO-spondin and low density lipoproteins from embryonic cerebrospinal fluid modulates their roles in early neurogenesis.Front Neuroanat. 2015 May 28;9:72. doi: 10.3389/fnana.2015.00072. eCollection 2015. Front Neuroanat. 2015. PMID: 26074785 Free PMC article.
-
SCO-spondin, a giant matricellular protein that regulates cerebrospinal fluid activity.Fluids Barriers CNS. 2021 Oct 2;18(1):45. doi: 10.1186/s12987-021-00277-w. Fluids Barriers CNS. 2021. PMID: 34600566 Free PMC article. Review.
-
Inhibition of thrombotic properties of persistent autoimmune anti-β2GPI antibodies in the mouse model of antiphospholipid syndrome.Blood. 2014 Feb 13;123(7):1090-7. doi: 10.1182/blood-2013-08-520882. Epub 2013 Nov 25. Blood. 2014. PMID: 24277078 Free PMC article.
-
Role of an intramolecular contact on lipoprotein uptake by the LDL receptor.Biochim Biophys Acta. 2011 Jun;1811(6):397-408. doi: 10.1016/j.bbalip.2011.04.002. Epub 2011 Apr 9. Biochim Biophys Acta. 2011. PMID: 21511053 Free PMC article.
-
Insight into Protein Engineering: From In silico Modelling to In vitro Synthesis.Curr Pharm Des. 2025;31(3):179-202. doi: 10.2174/0113816128349577240927071706. Curr Pharm Des. 2025. PMID: 39354773 Review.
References
-
- Brown, M. S., and Goldstein, J. L. (1986) Science 232 34–47 - PubMed
-
- Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W., and Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1 1–39 - PubMed
-
- Davis, C. G., Goldstein, J. L., Sudhof, T. C., Anderson, R. G., Russell, D. W., and Brown, M. S. (1987) Nature 326 760–765 - PubMed
-
- Lehrman, M. A., Goldstein, J. L., Brown, M. S., Russell, D. W., and Schneider, W. J. (1985) Cell 41 735–743 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

