A conformational switch in the DiGIR1 ribozyme involved in release and folding of the downstream I-DirI mRNA
- PMID: 19329537
- PMCID: PMC2673072
- DOI: 10.1261/rna.669209
A conformational switch in the DiGIR1 ribozyme involved in release and folding of the downstream I-DirI mRNA
Abstract
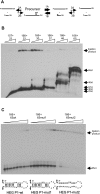
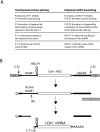
DiGIR1 is a group I-like cleavage ribozyme found as a structural domain within a nuclear twin-ribozyme group I intron. DiGIR1 catalyzes cleavage by branching at an Internal Processing Site (IPS) leading to formation of a lariat cap at the 5'-end of the 3'-cleavage product. The 3'-cleavage product is subsequently processed into an mRNA encoding a homing endonuclease. By analysis of combinations of 5'- and 3'-deletions, we identify a hairpin in the 5'-UTR of the mRNA (HEG P1) that is formed by conformational switching following cleavage. The formation of HEG P1 inhibits the reversal of the branching reaction, thus giving it directionality. Furthermore, the release of the mRNA is a consequence of branching rather than hydrolytic cleavage. A model is put forward that explains the release of the I-DirI mRNA with a lariat cap and a structured 5'-UTR as a direct consequence of the DiGIR1 branching reaction. The role of HEG P1 in GIR1 branching is reminiscent of that of hairpin P-1 in splicing of the Tetrahymena rRNA group I intron and illustrates a general principle in RNA-directed RNA processing.
Figures
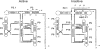
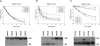



Similar articles
-
Intermolecular interaction between a branching ribozyme and associated homing endonuclease mRNA.Biol Chem. 2011 Apr;392(6):491-9. doi: 10.1515/BC.2011.055. Epub 2011 Apr 17. Biol Chem. 2011. PMID: 21495911
-
In vivo expression of the nucleolar group I intron-encoded I-dirI homing endonuclease involves the removal of a spliceosomal intron.EMBO J. 1999 Feb 15;18(4):1003-13. doi: 10.1093/emboj/18.4.1003. EMBO J. 1999. PMID: 10022842 Free PMC article.
-
The group I-like ribozyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis.Eur J Biochem. 2002 Dec;269(23):5804-12. doi: 10.1046/j.1432-1033.2002.03283.x. Eur J Biochem. 2002. PMID: 12444968
-
DiGIR1 and NaGIR1: naturally occurring group I-like ribozymes with unique core organization and evolved biological role.Biochimie. 2002 Sep;84(9):905-12. doi: 10.1016/s0300-9084(02)01443-8. Biochimie. 2002. PMID: 12458083 Review.
-
Structure and function of the hairpin ribozyme.J Mol Biol. 2000 Mar 24;297(2):269-91. doi: 10.1006/jmbi.2000.3560. J Mol Biol. 2000. PMID: 10715200 Review.
Cited by
-
Lariat capping as a tool to manipulate the 5' end of individual yeast mRNA species in vivo.RNA. 2017 May;23(5):683-695. doi: 10.1261/rna.059337.116. Epub 2017 Feb 3. RNA. 2017. PMID: 28159804 Free PMC article.
-
Convergent evolution of twintron-like configurations: One is never enough.RNA Biol. 2015;12(12):1275-88. doi: 10.1080/15476286.2015.1103427. RNA Biol. 2015. PMID: 26513606 Free PMC article. Review.
-
Speciation of a group I intron into a lariat capping ribozyme.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7659-64. doi: 10.1073/pnas.1322248111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821772 Free PMC article.
-
Nuclear group I introns in self-splicing and beyond.Mob DNA. 2013 Jun 5;4(1):17. doi: 10.1186/1759-8753-4-17. Mob DNA. 2013. PMID: 23738941 Free PMC article.
-
Molecular characterization of a new member of the lariat capping twin-ribozyme introns.Mob DNA. 2014 Sep 15;5:25. doi: 10.1186/1759-8753-5-25. eCollection 2014. Mob DNA. 2014. PMID: 25342998 Free PMC article.
References
-
- Agback P., Sandstrom A., Yamakage S., Sund C., Glemarec C., Chattopadhyaya J. Solution structure of lariat RNA by 500 MHz NMR spectroscopy and molecular dynamics studies in water. J. Biochem. Biophys. Methods. 1993;27:229–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
