In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes
- PMID: 19329564
- PMCID: PMC2675712
- DOI: 10.1104/pp.109.137703
In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes
Abstract
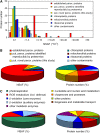
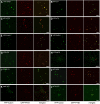
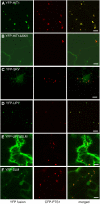
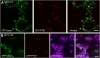
Peroxisomes are metabolically diverse organelles with essential roles in plant development. The major protein constituents of plant peroxisomes are well characterized, whereas only a few low-abundance and regulatory proteins have been reported to date. We performed an in-depth proteome analysis of Arabidopsis (Arabidopsis thaliana) leaf peroxisomes using one-dimensional gel electrophoresis followed by liquid chromatography and tandem mass spectrometry. We detected 65 established plant peroxisomal proteins, 30 proteins whose association with Arabidopsis peroxisomes had been previously demonstrated only by proteomic data, and 55 putative novel proteins of peroxisomes. We subsequently tested the subcellular targeting of yellow fluorescent protein fusions for selected proteins and confirmed the peroxisomal localization for 12 proteins containing predicted peroxisome targeting signals type 1 or 2 (PTS1/2), three proteins carrying PTS-related peptides, and four proteins that lack conventional targeting signals. We thereby established the tripeptides SLM> and SKV> (where > indicates the stop codon) as new PTS1s and the nonapeptide RVx(5)HF as a putative new PTS2. The 19 peroxisomal proteins conclusively identified from this study potentially carry out novel metabolic and regulatory functions of peroxisomes. Thus, this study represents an important step toward defining the complete plant peroxisomal proteome.
Figures




Comment in
-
In vivo subcellular targeting analysis validates a novel peroxisome targeting signal type 2 and the peroxisomal localization of two proteins with putative functions in defense in Arabidopsis.Plant Signal Behav. 2010 Feb;5(2):151-3. doi: 10.4161/psb.5.2.10412. Epub 2010 Feb 23. Plant Signal Behav. 2010. PMID: 20009535 Free PMC article.
References
-
- Aksam EB, Koek A, Kiel JA, Jourdan S, Veenhuis M, van der Klei IJ (2007) A peroxisomal Lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3 96–105 - PubMed
-
- Arai Y, Hayashi M, Nishimura M (2008) Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol 49 526–539 - PubMed
-
- Beevers H (1979) Microbodies in higher plants. Annu Rev Plant Physiol 30 159–193
-
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8 93–99
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

