Phosphate-responsive promoter of a Pichia pastoris sodium phosphate symporter
- PMID: 19329662
- PMCID: PMC2687300
- DOI: 10.1128/AEM.02913-08
Phosphate-responsive promoter of a Pichia pastoris sodium phosphate symporter
Abstract
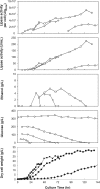
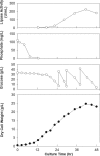
To develop a functional phosphate-regulated promoter in Pichia pastoris, a phosphate-responsive gene, PHO89, which encodes a putative sodium (Na(+))-coupled phosphate symporter, was isolated. Sequencing analyses revealed a 1,731-bp open reading frame encoding a 576-amino-acid polypeptide with 12 putative transmembrane domains. The properties of the PHO89 promoter (P(PHO89)) were investigated using a bacterial lipase gene as a reporter in 5-liter jar fermentation experiments. P(PHO89) was tightly regulated by phosphate and was highly activated when the cells were grown in a phosphate-limited external environment. Compared to translation elongation factor 1alpha and the glyceraldehyde-3-phosphate dehydrogenase promoter, P(PHO89) exhibited strong transcriptional activity with higher specific productivity (amount of lipase produced/cell/h). Furthermore, a cost-effective and simple P(PHO89)-based fermentation process was developed for industrial application. These results demonstrate the potential for efficient use of P(PHO89) for controlled production of recombinant proteins in P. pastoris.
Figures



References
-
- Ahn, J. O., E. S. Choi, H. W. Lee, S. H. Hwang, C. S. Kim, H. W. Jang, S. J. Haam, and J. K. Jung. 2004. Enhanced secretion of Bacillus stearothermophilus L1 lipase in Saccharomyces cerevisiae by translational fusion to cellulose-binding domain. Appl. Microbiol. Biotechnol. 64:833-839. - PubMed
-
- Ahn, J. O., J. Y. Hong, H.-W. Lee, M. S. Park, E. G. Lee, C. S. Kim, E. S. Choi, J. K. Jung, and H. W. Lee. 2007. Translation elongation factor 1-a gene from Pichia pastoris: molecular cloning, sequence, and use of its promoter. Appl. Microbiol. Biotechnol. 74:601-608. - PubMed
-
- Arocho, A., B. Chen, M. Ladanyi, and Q. Pan. 2006. Validation of the 2-ΔΔCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 15:56-61. - PubMed
-
- Bieche, I., M. Olivi, M. H. Champeme, D. Vidaud, R. Lidereau, and M. Vidaud. 1998. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int. J. Cancer 78:661-666. - PubMed
-
- Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. W. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

