DNA ligases: progress and prospects
- PMID: 19329793
- PMCID: PMC2719376
- DOI: 10.1074/jbc.R900017200
DNA ligases: progress and prospects
Abstract
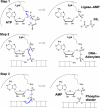
DNA ligases seal 5'-PO4 and 3'-OH polynucleotide ends via three nucleotidyl transfer steps involving ligase-adenylate and DNA-adenylate intermediates. DNA ligases are essential guardians of genomic integrity, and ligase dysfunction underlies human genetic disease syndromes. Crystal structures of DNA ligases bound to nucleotide and nucleic acid substrates have illuminated how ligase reaction chemistry is catalyzed, how ligases recognize damaged DNA ends, and how protein domain movements and active-site remodeling are used to choreograph the end-joining pathway. Although a shared feature of DNA ligases is their envelopment of the nicked duplex as a C-shaped protein clamp, they accomplish this feat by using remarkably different accessory structural modules and domain topologies. As structural, biochemical, and phylogenetic insights coalesce, we can expect advances on several fronts, including (i) pharmacological targeting of ligases for antibacterial and anticancer therapies and (ii) the discovery and design of new strand-sealing enzymes with unique substrate specificities.
Figures


Similar articles
-
DNA and RNA ligases: structural variations and shared mechanisms.Curr Opin Struct Biol. 2008 Feb;18(1):96-105. doi: 10.1016/j.sbi.2007.12.008. Epub 2008 Feb 8. Curr Opin Struct Biol. 2008. PMID: 18262407 Review.
-
Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.J Biol Chem. 2019 Mar 29;294(13):5094-5104. doi: 10.1074/jbc.RA119.007445. Epub 2019 Feb 4. J Biol Chem. 2019. PMID: 30718283 Free PMC article.
-
Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate.Mol Cell. 2007 Apr 27;26(2):257-71. doi: 10.1016/j.molcel.2007.02.026. Mol Cell. 2007. PMID: 17466627
-
Functional dissection of the DNA interface of the nucleotidyltransferase domain of chlorella virus DNA ligase.J Biol Chem. 2011 Apr 15;286(15):13314-26. doi: 10.1074/jbc.M111.226191. Epub 2011 Feb 18. J Biol Chem. 2011. PMID: 21335605 Free PMC article.
-
RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases.Mol Microbiol. 1995 Aug;17(3):405-10. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. Mol Microbiol. 1995. PMID: 8559059 Review.
Cited by
-
Biochemical and Structural Characterisation of DNA Ligases from Bacteria and Archaea.Biosci Rep. 2016 Oct 6;36(5):00391. doi: 10.1042/BSR20160003. Biosci Rep. 2016. PMID: 27582505 Free PMC article.
-
The expanding field of non-canonical RNA capping: new enzymes and mechanisms.R Soc Open Sci. 2021 May 19;8(5):201979. doi: 10.1098/rsos.201979. R Soc Open Sci. 2021. PMID: 34017598 Free PMC article. Review.
-
Fragment-based discovery of 6-azaindazoles as inhibitors of bacterial DNA ligase.ACS Med Chem Lett. 2013 Oct 18;4(12):1208-12. doi: 10.1021/ml4003277. eCollection 2013 Dec 12. ACS Med Chem Lett. 2013. PMID: 24900632 Free PMC article.
-
From Structure-Function Analyses to Protein Engineering for Practical Applications of DNA Ligase.Archaea. 2015 Oct 5;2015:267570. doi: 10.1155/2015/267570. eCollection 2015. Archaea. 2015. PMID: 26508902 Free PMC article. Review.
-
A high-throughput assay for the comprehensive profiling of DNA ligase fidelity.Nucleic Acids Res. 2016 Jan 29;44(2):e14. doi: 10.1093/nar/gkv898. Epub 2015 Sep 13. Nucleic Acids Res. 2016. PMID: 26365241 Free PMC article.
References
-
- Lehman I. R. ( 1974) Science 186, 790– 797 - PubMed
-
- Tomkinson A. E., Vijayakumar S., Pascal J. M., Ellenberger T. ( 2006) Chem. Rev. 106, 687– 699 - PubMed
-
- Shuman S., Glickman M. S. ( 2007) Nat. Rev. Microbiol. 5, 852– 861 - PubMed
-
- Odell M., Sriskanda V., Shuman S., Nikolov D. ( 2000) Mol. Cell 6, 1183– 1193 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

