Solution structure of a novel T-cell adhesion inhibitor derived from the fragment of ICAM-1 receptor: cyclo(1,8)-Cys-Pro-Arg-Gly-Gly-Ser-Val-Cys
- PMID: 19330816
- PMCID: PMC2742958
- DOI: 10.1002/bip.21192
Solution structure of a novel T-cell adhesion inhibitor derived from the fragment of ICAM-1 receptor: cyclo(1,8)-Cys-Pro-Arg-Gly-Gly-Ser-Val-Cys
Abstract
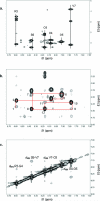
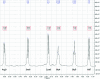
This study is aimed at elucidating the structure of a novel T-cell adhesion inhibitor, cyclo(1,8)-CPRGGSVC using one- and two-dimensional (2D) (1)H NMR and molecular dynamics (MD) simulation. The peptide is derived from the sequence of its parent peptide cIBR (cyclo(1,12)-PenPRGGSVLVTGC), which is a fragment of intercellular adhesion molecule-1 (ICAM-1). Our previous results show that the cyclo(1,8)-CPRGGSVC peptide binds to the LFA-1 I-domain and inhibits heterotypic T-cell adhesion, presumably by blocking the LFA-1/ICAM-1 interactions. The structure of the peptide was determined using NMR and MD simulation in aqueous solution. Our results indicate that the peptide adopts type-I beta-turn conformation at the Pro2-Arg3-Gly4-Gly5 (PRGG) sequence. The beta-turn structure at the PRGG motif is well conserved in cIBR peptide and ICAM-1 receptor, which suggests the importance of the PRGG motif for the biological activity of cyclo(1,8)-CPRGGSVC peptide. Meanwhile, the Gly5-Ser6-Val7-Cys8-Cys1 (GSVCC) sequence forms a "turn-like" random coil structure that does not belong to any structured motif. Therefore, cyclo(1,8)-CPRGGSVC peptide has only one structured region at the PRGG sequence, which may play an important role in the binding of the peptide to the LFA-1 I-domain. The conserved beta-turn conformation of the PRGG motif in ICAM-1, cIBR, and cyclo(1,8)-CPRGGSVC peptides can potentially be used to design peptidomimetics. (c) 2009 Wiley Periodicals, Inc. Biopolymers 91: 633-641, 2009.This article was originally published online as an accepted preprint. The "Published Online" date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com.
Figures



Similar articles
-
Structural recognition of an ICAM-1 peptide by its receptor on the surface of T cells: conformational studies of cyclo (1, 12)-Pen-Pro-Arg-Gly-Gly-Ser-Val-Leu-Val-Thr-Gly-Cys-OH.J Pept Res. 1999 Apr;53(4):422-31. doi: 10.1034/j.1399-3011.1999.00080.x. J Pept Res. 1999. PMID: 10406220
-
Structural modifications of ICAM-1 cyclic peptides to improve the activity to inhibit heterotypic adhesion of T cells.Chem Biol Drug Des. 2008 Jul;72(1):27-33. doi: 10.1111/j.1747-0285.2008.00676.x. Epub 2008 Jun 11. Chem Biol Drug Des. 2008. PMID: 18554252 Free PMC article.
-
Structural and ICAM-1-docking properties of a cyclic peptide from the I-domain of LFA-1: an inhibitor of ICAM-1/LFA- 1-mediated T-cell adhesion.J Biomol Struct Dyn. 2002 Apr;19(5):789-99. doi: 10.1080/07391102.2002.10506785. J Biomol Struct Dyn. 2002. PMID: 11922836
-
Targeting ICAM-1/LFA-1 interaction for controlling autoimmune diseases: designing peptide and small molecule inhibitors.Peptides. 2003 Mar;24(3):487-501. doi: 10.1016/s0196-9781(03)00083-4. Peptides. 2003. PMID: 12732350 Review.
-
Review: rhinoviruses and their ICAM receptors.J Struct Biol. 1999 Dec 1;128(1):69-74. doi: 10.1006/jsbi.1999.4143. J Struct Biol. 1999. PMID: 10600561 Review.
References
-
- Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. Annu Rev Immunol. 2001;19:375–396. - PubMed
-
- Davis DM, Dustin ML. Trends Immunol. 2004;25:323–327. - PubMed
-
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. Science. 1999;285:221–227. - PubMed
-
- Bromley SK, Iaboni A, Davis SJ, Whitty A, Green JM, Shaw AS, Weiss A, Dustin ML. Nat Immunol. 2001;2:1159–1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

