The homeodomain protein Cux1 interacts with Grg4 to repress p27 kip1 expression during kidney development
- PMID: 19332113
- PMCID: PMC2742960
- DOI: 10.1016/j.gene.2009.03.014
The homeodomain protein Cux1 interacts with Grg4 to repress p27 kip1 expression during kidney development
Abstract
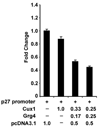
The homeodomain protein Cux1 is highly expressed in the nephrogenic zone of the developing kidney where it functions to regulate cell proliferation. Here we show that Cux1 directly interacts with the co-repressor Grg4 (Groucho 4), a known effector of Notch signaling. Promoter reporter based luciferase assays revealed enhanced repression of p27(kip1) promoter activity by Cux1 in the presence of Grg4. Chromatin immunoprecipitation (ChIP) assays demonstrated the direct interaction of Cux1 with p27(kip1) in newborn kidney tissue in vivo. ChIP assays also identified interactions of Cux1, Grg4, HDAC1, and HDAC3 with p27(kip1) at two separate sites in the p27(kip1) promoter. DNAse1 footprinting experiments revealed that Cux1 binds to the p27(kip1) promoter on the sequence containing two Sp1 sites and a CCAAT box approximately 500 bp from the transcriptional start site, and to an AT rich sequence approximately 1.5 kb from the transcriptional start site. Taken together, these results identify Grg4 as an interacting partner for Cux1 and suggest a mechanism of p27(kip1) repression by Cux1 during kidney development.
Figures



Similar articles
-
Cux1 promotes cell proliferation and polycystic kidney disease progression in an ADPKD mouse model.Am J Physiol Renal Physiol. 2017 Oct 1;313(4):F1050-F1059. doi: 10.1152/ajprenal.00380.2016. Epub 2017 Jul 12. Am J Physiol Renal Physiol. 2017. PMID: 28701314 Free PMC article.
-
Cux1 regulation of the cyclin kinase inhibitor p27kip1 in polycystic kidney disease is attenuated by HDAC inhibitors.Gene. 2019;721S:100007. doi: 10.1016/j.gene.2019.100007. Epub 2019 Feb 12. Gene. 2019. PMID: 34531000
-
Cux1 regulation of the cyclin kinase inhibitor p27kip1 in polycystic kidney disease is attenuated by HDAC inhibitors.Gene X. 2019 Jun;2:100007. doi: 10.1016/j.gene.2019.100007. Epub 2019 Feb 12. Gene X. 2019. PMID: 31396588 Free PMC article.
-
The multiple roles of CUX1: insights from mouse models and cell-based assays.Gene. 2008 Apr 15;412(1-2):84-94. doi: 10.1016/j.gene.2008.01.017. Epub 2008 Feb 2. Gene. 2008. PMID: 18313863 Review.
-
CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers.Nat Rev Cancer. 2014 Oct;14(10):673-82. doi: 10.1038/nrc3805. Epub 2014 Sep 5. Nat Rev Cancer. 2014. PMID: 25190083 Review.
Cited by
-
Regulation of retinoic acid synthetic enzymes by WT1 and HDAC inhibitors in 293 cells.Int J Mol Med. 2017 Sep;40(3):661-672. doi: 10.3892/ijmm.2017.3051. Epub 2017 Jul 3. Int J Mol Med. 2017. PMID: 28677722 Free PMC article.
-
The Groucho/Transducin-like enhancer of split protein family in animal development.IUBMB Life. 2015 Jul;67(7):472-81. doi: 10.1002/iub.1395. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26172616 Free PMC article. Review.
-
The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons.PLoS One. 2010 May 11;5(5):e10596. doi: 10.1371/journal.pone.0010596. PLoS One. 2010. PMID: 20485671 Free PMC article.
-
Cux1 promotes cell proliferation and polycystic kidney disease progression in an ADPKD mouse model.Am J Physiol Renal Physiol. 2017 Oct 1;313(4):F1050-F1059. doi: 10.1152/ajprenal.00380.2016. Epub 2017 Jul 12. Am J Physiol Renal Physiol. 2017. PMID: 28701314 Free PMC article.
-
Inhibition of Notch pathway attenuates the progression of human immunodeficiency virus-associated nephropathy.Am J Physiol Renal Physiol. 2013 Apr 15;304(8):F1127-36. doi: 10.1152/ajprenal.00475.2012. Epub 2013 Feb 6. Am J Physiol Renal Physiol. 2013. PMID: 23389453 Free PMC article.
References
-
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. - PubMed
-
- Sansregret L, Nepveu A. The multiple roles of Cux1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. - PubMed
-
- Gupta S, Luong MX, Bleuming SA, Miele A, Luong M, Young D, Knudsen ES, Van Wijnen AJ, Stein JL, Stein GS. Tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J Cell Physiol. 2003;196:541–556. - PubMed
-
- Lievens PM, Donady JJ, Tufarelli C, Neufeld EJ. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

