Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy
- PMID: 19332114
- PMCID: PMC4152850
- DOI: 10.1016/j.ygeno.2009.03.003
Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy
Abstract
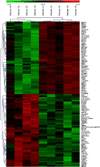
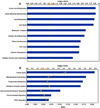
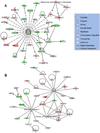
We employed ABI high-density oligonucleotide microarrays containing 31,700 sixty-mer probes (representing 27,868 annotated human genes) to determine differential gene expression in idiopathic dilated cardiomyopathy (DCM). We identified 626 up-regulated and 636 down-regulated genes in DCM compared to controls. Most significant changes occurred in the tricarboxylic acid cycle, angiogenesis, and apoptotic signaling pathways, among which 32 apoptosis- and 13 MAPK activity-related genes were altered. Inorganic cation transporter, catalytic activities, energy metabolism and electron transport-related processes were among the most critically influenced pathways. Among the up-regulated genes were HTRA1 (6.9-fold), PDCD8(AIFM1) (5.2) and PRDX2 (4.4) and the down-regulated genes were NR4A2 (4.8), MX1 (4.3), LGALS9 (4), IFNA13 (4), UNC5D (3.6) and HDAC2 (3) (p<0.05), all of which have no clearly defined cardiac-related function yet. Gene ontology and enrichment analysis also revealed significant alterations in mitochondrial oxidative phosphorylation, metabolism and Alzheimer's disease pathways. Concordance was also confirmed for a significant number of genes and pathways in an independent validation microarray dataset. Furthermore, verification by real-time RT-PCR showed a high degree of consistency with the microarray results. Our data demonstrate an association of DCM with alterations in various cellular events and multiple yet undeciphered genes that may contribute to heart muscle disease pathways.
Figures

Similar articles
-
Integrated Left Ventricular Global Transcriptome and Proteome Profiling in Human End-Stage Dilated Cardiomyopathy.PLoS One. 2016 Oct 6;11(10):e0162669. doi: 10.1371/journal.pone.0162669. eCollection 2016. PLoS One. 2016. PMID: 27711126 Free PMC article.
-
Global gene expression in human myocardium-oligonucleotide microarray analysis of regional diversity and transcriptional regulation in heart failure.J Mol Med (Berl). 2004 May;82(5):308-16. doi: 10.1007/s00109-004-0527-2. Epub 2004 Apr 23. J Mol Med (Berl). 2004. PMID: 15103417
-
Gene expression profiles in end-stage human idiopathic dilated cardiomyopathy: altered expression of apoptotic and cytoskeletal genes.Genomics. 2004 Feb;83(2):281-97. doi: 10.1016/j.ygeno.2003.08.007. Genomics. 2004. PMID: 14706457
-
Inflammatory dilated cardiomyopathy (DCMI).Herz. 2005 Sep;30(6):535-44. doi: 10.1007/s00059-005-2730-5. Herz. 2005. PMID: 16170686 Review.
-
Global gene expression profiling in the failing myocardium.Circ J. 2009 Sep;73(9):1568-76. doi: 10.1253/circj.cj-09-0465. Epub 2009 Jul 29. Circ J. 2009. PMID: 19638707 Review.
Cited by
-
Plasma levels of alpha-1-antichymotrypsin are elevated in patients with chronic heart failure, but are of limited prognostic value.Neth Heart J. 2014 Sep;22(9):391-5. doi: 10.1007/s12471-014-0584-2. Neth Heart J. 2014. PMID: 25172361 Free PMC article.
-
Mechano-growth factor E-domain modulates cardiac contractile function through 14-3-3 protein interactomes.Front Physiol. 2022 Nov 16;13:1028345. doi: 10.3389/fphys.2022.1028345. eCollection 2022. Front Physiol. 2022. PMID: 36467694 Free PMC article.
-
Cardiac-specific deletion of the microtubule-binding protein CENP-F causes dilated cardiomyopathy.Dis Model Mech. 2012 Jul;5(4):468-80. doi: 10.1242/dmm.008680. Epub 2012 Mar 22. Dis Model Mech. 2012. PMID: 22563055 Free PMC article.
-
The histone 3 lysine 9 methyltransferase inhibitor chaetocin improves prognosis in a rat model of high salt diet-induced heart failure.Sci Rep. 2017 Jan 4;7:39752. doi: 10.1038/srep39752. Sci Rep. 2017. PMID: 28051130 Free PMC article.
-
2-[18F]Fluoropropionic Acid PET Imaging of Doxorubicin-Induced Cardiotoxicity.Mol Imaging Biol. 2025 Feb;27(1):109-119. doi: 10.1007/s11307-024-01978-y. Epub 2025 Jan 14. Mol Imaging Biol. 2025. PMID: 39810069 Free PMC article.
References
-
- Ross J., Jr. Dilated cardiomyopathy: concepts derived from gene deficient and transgenic animal models. Circ. J. 2002;66(3):219–224. - PubMed
-
- Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail. Rev. 2005;10(3):225–235. - PubMed
-
- Granzier H, Wu Y, Siegfried L, LeWinter M. Titin: physiological function and role in cardiomyopathy and failure. Heart Fail. Rev. 2005;10(3):211–223. - PubMed
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu. Rev. Genomics Hum. Genet. 2005;6:185–216. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

