The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3)
- PMID: 19332543
- PMCID: PMC2682855
- DOI: 10.1074/jbc.M900370200
The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3)
Abstract
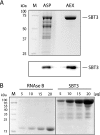
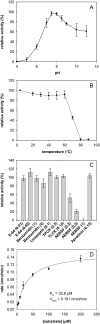
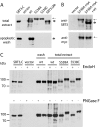
A transgenic plant cell suspension culture was established as a versatile and efficient expression system for the subtilase SlSBT3 from tomato. The recombinant protease was purified to homogeneity from culture supernatants by fractionated ammonium sulfate precipitation, batch adsorption to cation exchange material, and anion exchange chromatography. Purified SlSBT3 was identified as a 79-kDa glycoprotein with both complex and paucimannosidic type glycan chains at Asn(177), Asn(203), Asn(376), Asn(697), and Asn(745). SlSBT3 was found to be a very stable enzyme, being fully active at 60 degrees C and showing highest activity at alkaline conditions with a maximum between pH 7.5 and 8.0. Substrate specificity of SlSBT3 was analyzed in detail, revealing a preference for Gln and Lys in the P(1) and P(2) positions of oligopeptide substrates, respectively. Similar to bacterial, yeast, and mammalian subtilases, SlSBT3 is synthesized as a preproenzyme, and processing of the prodomain in the endoplasmic reticulum is a prerequisite for passage through the secretory pathway. SlSBT3 S538A and S538C active site mutants accumulated intracellularly as unprocessed zymogens, indicating that prodomain cleavage occurs autocatalytically. The wild-type SlSBT3 protein failed to cleave the prodomain of the S538A mutant in trans, demonstrating that zymogen maturation is an intramolecular process. Distinguishing features of plant as compared with mammalian subtilases include the insertion of a large protease-associated domain between the His and Ser residues of the catalytic triad and the C-terminal extension to the catalytic domain. Both features were found to be required for SlSBT3 activity and, consequently, for prodomain processing and secretion.
Figures



References
-
- Dodson, G., and Wlodawer, A. (1998) Trends Biochem. Sci. 23 347–352 - PubMed
-
- Seidah, N. G., Khatib, A. M., and Prat, A. (2006) Biol. Chem. 387 871–877 - PubMed
-
- Sakai, J., Rawson, R. B., Espenshade, P. J., Cheng, D., Seegmiller, A. C., Goldstein, J. L., and Brown, M. S. (1998) Mol. Cell 2 505–514 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

