Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries
- PMID: 19332564
- PMCID: PMC2682014
- DOI: 10.1128/MCB.01655-08
Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries
Abstract
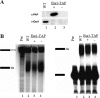
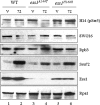
The C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) is a reiterated heptad sequence (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) that plays a key role in the transcription cycle, coordinating the exchange of transcription and RNA processing factors. The structure of the CTD is flexible and undergoes conformational changes in response to serine phosphorylation and proline isomerization. Here we report that the Ess1 peptidyl prolyl isomerase functionally interacts with the transcription initiation factor TFIIB and with the Ssu72 CTD phosphatase and Pta1 components of the CPF 3'-end processing complex. The ess1(A144T) and ess1(H164R) mutants, initially described by Hanes and coworkers (Yeast 5:55-72, 1989), accumulate the pSer5 phosphorylated form of Pol II; confer phosphate, galactose, and inositol auxotrophies; and fail to activate PHO5, GAL10, and INO1 reporter genes. These mutants are also defective for transcription termination, but in vitro experiments indicate that this defect is not caused by altering the processing efficiency of the cleavage/polyadenylation machinery. Consistent with a role in initiation and termination, Ess1 associates with the promoter and terminator regions of the PMA1 and PHO5 genes. We propose that Ess1 facilitates pSer5-Pro6 dephosphorylation by generating the CTD structural conformation recognized by the Ssu72 phosphatase and that pSer5 dephosphorylation affects both early and late stages of the transcription cycle.
Figures




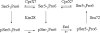
Similar articles
-
Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase II transcription cycle.Mol Cell Biol. 2012 Sep;32(17):3594-607. doi: 10.1128/MCB.00672-12. Epub 2012 Jul 9. Mol Cell Biol. 2012. PMID: 22778132 Free PMC article.
-
The Ess1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle.Biochim Biophys Acta. 2014;1839(4):316-33. doi: 10.1016/j.bbagrm.2014.02.001. Epub 2014 Feb 12. Biochim Biophys Acta. 2014. PMID: 24530645 Free PMC article. Review.
-
The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway.Mol Cell. 2009 Oct 23;36(2):255-66. doi: 10.1016/j.molcel.2009.08.018. Mol Cell. 2009. PMID: 19854134 Free PMC article.
-
The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae.Genetics. 2003 Dec;165(4):1687-702. doi: 10.1093/genetics/165.4.1687. Genetics. 2003. PMID: 14704159 Free PMC article.
-
Diverse and conserved roles of the protein Ssu72 in eukaryotes: from yeast to higher organisms.Curr Genet. 2021 Apr;67(2):195-206. doi: 10.1007/s00294-020-01132-5. Epub 2020 Nov 26. Curr Genet. 2021. PMID: 33244642 Review.
Cited by
-
Domain acquisition enabled functional expansion of the TFIIS transcription factor family.Cell Biosci. 2025 Jun 4;15(1):78. doi: 10.1186/s13578-025-01423-9. Cell Biosci. 2025. PMID: 40468406 Free PMC article. Review.
-
The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination.Mol Cell Biol. 2012 Nov;32(21):4306-22. doi: 10.1128/MCB.00382-12. Epub 2012 Aug 20. Mol Cell Biol. 2012. PMID: 22907757 Free PMC article.
-
SPOC domain proteins in health and disease.Genes Dev. 2023 Mar 1;37(5-6):140-170. doi: 10.1101/gad.350314.122. Epub 2023 Mar 16. Genes Dev. 2023. PMID: 36927757 Free PMC article. Review.
-
TAL effectors target the C-terminal domain of RNA polymerase II (CTD) by inhibiting the prolyl-isomerase activity of a CTD-associated cyclophilin.PLoS One. 2012;7(7):e41553. doi: 10.1371/journal.pone.0041553. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911812 Free PMC article.
-
RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts.PLoS One. 2012;7(6):e38769. doi: 10.1371/journal.pone.0038769. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701709 Free PMC article.
References
-
- Arevalo-Rodriguez, M., X. Wu, S. D. Hanes, and J. Heitman. 2004. Prolyl isomerases in yeast. Front. Biosci. 92420-2446. - PubMed
-
- Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. - PubMed
-
- Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17257-261. - PubMed
-
- Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 71013-1023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
