The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae
- PMID: 19332891
- PMCID: PMC2700514
- DOI: 10.1083/jcb.200811119
The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae
Abstract
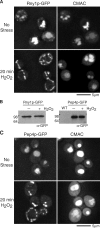
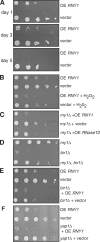
The cellular response to stress conditions involves a decision between survival or cell death when damage is severe. A conserved stress response in eukaryotes involves endonucleolytic cleavage of transfer RNAs (tRNAs). The mechanism and significance of such tRNA cleavage is unknown. We show that in yeast, tRNAs are cleaved by the RNase T2 family member Rny1p, which is released from the vacuole into the cytosol during oxidative stress. Rny1p modulates yeast cell survival during oxidative stress independently of its catalytic ability. This suggests that upon release to the cytosol, Rny1p promotes cell death by direct interactions with downstream components. Thus, detection of Rny1p, and possibly its orthologues, in the cytosol may be a conserved mechanism for assessing cellular damage and determining cell survival, analogous to the role of cytochrome c as a marker for mitochondrial damage.
Figures



References
-
- Acquati F., Nucci C., Bianchi M.G., Gorletta T., Taramelli R. 2001. Molecular cloning, tissue distribution, and chromosomal localization of the human homolog of the R2/Th/Stylar ribonuclease gene family.Methods Mol. Biol. 160:87–101 - PubMed
-
- Borek E., Baliga B.S., Gehrke C.W., Kuo C.W., Belman S., Troll W., Waalkes T.P. 1977. High turnover rate of transfer RNA in tumor tissue.Cancer Res. 37:3362–3366 - PubMed
-
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors.Gene. 110:119–122 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

