Characterisation of the fibroblast growth factor dependent transcriptome in early development
- PMID: 19333377
- PMCID: PMC2659300
- DOI: 10.1371/journal.pone.0004951
Characterisation of the fibroblast growth factor dependent transcriptome in early development
Abstract
Background: FGF signaling has multiple roles in regulating processes in animal development, including the specification and patterning of the mesoderm. In addition, FGF signaling supports self renewal of human embryonic stem cells and is required for differentiation of murine embryonic stem cells into a number of lineages.
Methodology/principal findings: Given the importance of FGF signaling in regulating development and stem cell behaviour, we aimed to identify the transcriptional targets of FGF signalling during early development in the vertebrate model Xenopus laevis. We analysed the effects on gene expression in embryos in which FGF signaling was inhibited by dominant negative FGF receptors. 67 genes positively regulated by FGF signaling and 16 genes negatively regulated by FGF signaling were identified. FGF target genes are expressed in distinct waves during the late blastula to early gastrula phase. Many of these genes are expressed in the early mesoderm and dorsal ectoderm. A widespread requirement for FGF in regulating genes expressed in the Spemann organizer is revealed. The FGF targets MKP1 and DUSP5 are shown to be negative regulators of FGF signaling in early Xenopus tissues. FoxD3 and Lin28, which are involved in regulating pluripotency in ES cells are shown to be down regulated when FGF signaling is blocked.
Conclusions: We have undertaken a detailed analysis of FGF target genes which has generated a robust, well validated data set. We have found a widespread role for FGF signaling in regulating the expression of genes mediating the function of the Spemann organizer. In addition, we have found that the FGF targets MKP1 and DUSP5 are likely to contribute to the complex feedback loops involved in modulating responses to FGF signaling. We also find a link between FGF signaling and the expression of known regulators of pluripotency.
Conflict of interest statement
Figures
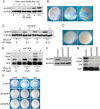

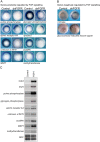

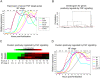

References
-
- Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. - PubMed
-
- Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. - PubMed
-
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. - PubMed
-
- Mathieu J, Griffin K, Herbomel P, Dickmeis T, Strahle U, et al. Nodal and Fgf pathways interact through a positive regulatory loop and synergize to maintain mesodermal cell populations. Development. 2004;131:629–641. - PubMed
-
- Amaya E, Stein PA, Musci TJ, Kirschner MW. FGF signalling in the early specification of mesoderm in Xenopus. development. 1993;118:477–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

