A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system
- PMID: 19333389
- PMCID: PMC2659438
- DOI: 10.1371/journal.pone.0005070
A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system
Abstract
Background: 4E-BP is a translational inhibitor that binds to eIF4E to repress cap-dependent translation initiation. This critical protein:protein interaction is regulated by the phosphorylation of 4E-BP. Hypophosphorylated 4E-BP binds to eIF4E and inhibits cap-dependent translation, whereas hyperphosphorylated forms do not. While three 4E-BP proteins exist in mammals, only one gene encoding for 4E-BP is present in the sea urchin genome. The protein product has a highly conserved core domain containing the eIF4E-binding domain motif (YxxxxLPhi) and four of the regulatory phosphorylation sites.
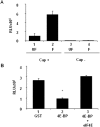
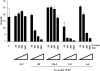
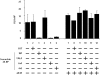
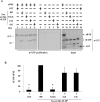
Methodology/principal findings: Using a sea urchin cell-free cap-dependent translation system prepared from fertilized eggs, we provide the first direct evidence that the sea urchin 4E-BP inhibits cap-dependent translation. We show here that a sea urchin 4E-BP variant, mimicking phosphorylation on four core residues required to abrogate binding to eIF4E, surprisingly maintains physical association to eIF4E and inhibits protein synthesis.
Conclusions/significance: Here, we examine the involvement of the evolutionarily conserved core domain and phosphorylation sites of sea urchin 4E-BP in the regulation of eIF4E-binding. These studies primarily demonstrate the conserved activity of the 4E-BP translational repressor and the importance of the eIF4E-binding domain in sea urchin. We also show that a variant mimicking hyperphosphorylation of the four regulatory phosphorylation sites common to sea urchin and human 4E-BP is not sufficient for release from eIF4E and translation promotion. Therefore, our results suggest that there are additional mechanisms to that of phosphorylation at the four critical sites of 4E-BP that are required to disrupt binding to eIF4E.
Conflict of interest statement
Figures



References
-
- Mathews MB, Sonenberg N, Hershey JWB. Origins and Principles of Translational Control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007.
-
- Davidson EH, Hough-Evans BR, Britten RJ. Molecular biology of the sea urchin embryo. Science. 1982;217:17–26. - PubMed
-
- Epel D. The initiation of development at fertilization. Cell Differ Dev. 1990;29:1–12. - PubMed
-
- Humphreys T. Measurements of messenger RNA entering polysomes upon fertilization of sea urchin eggs. Dev Biol. 1971;26:201–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

