Length variations amongst protein domain superfamilies and consequences on structure and function
- PMID: 19333395
- PMCID: PMC2659687
- DOI: 10.1371/journal.pone.0004981
Length variations amongst protein domain superfamilies and consequences on structure and function
Abstract
Background: Related protein domains of a superfamily can be specified by proteins of diverse lengths. The structural and functional implications of indels in a domain scaffold have been examined.
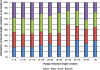
Methodology: In this study, domain superfamilies with large length variations (more than 30% difference from average domain size, referred as 'length-deviant' superfamilies and 'length-rigid' domain superfamilies (<10% length difference from average domain size) were analyzed for the functional impact of such structural differences. Our delineated dataset, derived from an objective algorithm, enables us to address indel roles in the presence of peculiar structural repeats, functional variation, protein-protein interactions and to examine 'domain contexts' of proteins tolerant to large length variations. Amongst the top-10 length-deviant superfamilies analyzed, we found that 80% of length-deviant superfamilies possess distant internal structural repeats and nearly half of them acquired diverse biological functions. In general, length-deviant superfamilies have higher chance, than length-rigid superfamilies, to be engaged in internal structural repeats. We also found that approximately 40% of length-deviant domains exist as multi-domain proteins involving interactions with domains from the same or other superfamilies. Indels, in diverse domain superfamilies, were found to participate in the accretion of structural and functional features amongst related domains. With specific examples, we discuss how indels are involved directly or indirectly in the generation of oligomerization interfaces, introduction of substrate specificity, regulation of protein function and stability.
Conclusions: Our data suggests a multitude of roles for indels that are specialized for domain members of different domain superfamilies. These specialist roles that we observe and trends in the extent of length variation could influence decision making in modeling of new superfamily members. Likewise, the observed limits of length variation, specific for each domain superfamily would be particularly relevant in the choice of alignment length search filters commonly applied in protein sequence analysis.
Conflict of interest statement
Figures
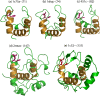




References
-
- Heringa J, Taylor WR. Three-dimensional domain duplication, swapping and stealing. Curr Opin Struct Biol. 1997;7:416–421. - PubMed
-
- Heringa J. Detection of internal repeats: how common are they? Curr Opin Struct Biol. 1998;8:338–345. - PubMed
-
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. - PubMed
-
- Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. - PubMed
-
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

