Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors
- PMID: 19333550
- PMCID: PMC11115575
- DOI: 10.1007/s00018-009-0020-8
Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors
Abstract
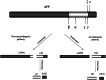
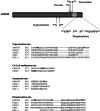
The Alzheimer's amyloid precursor protein (APP) belongs to a conserved gene family that also includes the mammalian APLP1 and APLP2, the Drosophila APPL, and the C. elegans APL-1. The biological function of APP is still not fully clear. However, it is known that the APP family proteins have redundant and partly overlapping functions, which demonstrates the importance of studying all APP family members to gain a more complete picture. When APP was first cloned, it was speculated that it could function as a receptor. This theory has been further substantiated by studies showing that APP and its homologues bind both extracellular ligands and intracellular adaptor proteins. The APP family proteins undergo regulated intramembrane proteolysis (RIP), generating secreted and cytoplasmic fragments that have been ascribed different functions. In this review, we will discuss the APP family with focus on biological functions, binding partners, and regulated processing.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

