Chromatin immunoprecipitation in early Xenopus laevis embryos
- PMID: 19334278
- PMCID: PMC2832845
- DOI: 10.1002/dvdy.21931
Chromatin immunoprecipitation in early Xenopus laevis embryos
Abstract
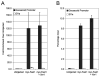
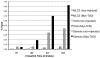
Chromatin immunoprecipitation (ChIP) is a powerful method for analyzing the interaction of regulatory proteins with genomic loci, but has been difficult to apply to studies on early embryos due to the limiting amount of genomic material in these samples. Here, we present a comprehensive technique for performing ChIP on blastula and gastrula stage Xenopus embryos. We also describe methods for optimizing crosslinking and chromatin shearing, verifying antibody specificity, maximizing PCR sensitivity, and quantifying PCR results, allowing for the use of as few as 50 early blastula stage embryos (approximately 5x10(4) cells) per experimental condition. Finally, we demonstrate the predicted binding of endogenous beta-catenin to the nodal-related 6 promoter, binding of tagged Fast-1/FoxH1 to the goosecoid promoter, and binding of tagged Tcf3 to the siamois and nodal-related 6 promoters as examples of the potential application of ChIP to embryological investigations. Developmental Dynamics 238:1422-1432, 2009. (c) 2009 Wiley-Liss, Inc.
Figures



References
-
- Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008:1032–1045. - PubMed
-
- Green DM, Sessions SK. Amphibian cytogenetics and evolution. xv. San Diego: Academic Press; 1991. p. 456.
-
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000:124–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

