Genic non-coding microsatellites in the rice genome: characterization, marker design and use in assessing genetic and evolutionary relationships among domesticated groups
- PMID: 19335879
- PMCID: PMC2680414
- DOI: 10.1186/1471-2164-10-140
Genic non-coding microsatellites in the rice genome: characterization, marker design and use in assessing genetic and evolutionary relationships among domesticated groups
Abstract
Background: Completely sequenced plant genomes provide scope for designing a large number of microsatellite markers, which are useful in various aspects of crop breeding and genetic analysis. With the objective of developing genic but non-coding microsatellite (GNMS) markers for the rice (Oryza sativa L.) genome, we characterized the frequency and relative distribution of microsatellite repeat-motifs in 18,935 predicted protein coding genes including 14,308 putative promoter sequences.
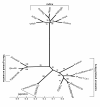
Results: We identified 19,555 perfect GNMS repeats with densities ranging from 306.7/Mb in chromosome 1 to 450/Mb in chromosome 12 with an average of 357.5 GNMS per Mb. The average microsatellite density was maximum in the 5' untranslated regions (UTRs) followed by those in introns, promoters, 3'UTRs and minimum in the coding sequences (CDS). Primers were designed for 17,966 (92%) GNMS repeats, including 4,288 (94%) hypervariable class I types, which were bin-mapped on the rice genome. The GNMS markers were most polymorphic in the intronic region (73.3%) followed by markers in the promoter region (53.3%) and least in the CDS (26.6%). The robust polymerase chain reaction (PCR) amplification efficiency and high polymorphic potential of GNMS markers over genic coding and random genomic microsatellite markers suggest their immediate use in efficient genotyping applications in rice. A set of these markers could assess genetic diversity and establish phylogenetic relationships among domesticated rice cultivar groups. We also demonstrated the usefulness of orthologous and paralogous conserved non-coding microsatellite (CNMS) markers, identified in the putative rice promoter sequences, for comparative physical mapping and understanding of evolutionary and gene regulatory complexities among rice and other members of the grass family. The divergence between long-grained aromatics and subspecies japonica was estimated to be more recent (0.004 Mya) compared to short-grained aromatics from japonica (0.006 Mya) and long-grained aromatics from subspecies indica (0.014 Mya).
Conclusion: Our analyses showed that GNMS markers with their high polymorphic potential would be preferred candidate functional markers in various marker-based applications in rice genetics, genomics and breeding. The CNMS markers provided encouraging implications for their use in comparative genome mapping and understanding of evolutionary complexities in rice and other members of grass family.
Figures
References
-
- Hancock JM. Microsatellites and other simple sequences: genomic context and mutational mechanisms. In: Goldstein DB, Schlotterer C, editor. Microsatellite: evolution and applications. Oxford University Press, Oxford, U.K.; 1999. pp. 1–9.
-
- Kim GP, Colangelo L, Allegra C, Glebov O, Parr A, Hooper S, Williams J, Paik SM, Eaton L, King W, Wolmark N, Wieand HS, Ilan R. Prognostic role of microsatellite instability in colon cancer. Proc Am Soc Clin Oncol. 2001;20:1666.
-
- Streelman JT, Kocher D. Microsatellite variation associated with prolactin expression and growth of salt challenged Tilapia. Physiol Genomics. 2002;9:1–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

