Real-time monitoring of aRNA production during T7 amplification to prevent the loss of sample representation during microarray hybridization sample preparation
- PMID: 19336411
- PMCID: PMC2677895
- DOI: 10.1093/nar/gkp193
Real-time monitoring of aRNA production during T7 amplification to prevent the loss of sample representation during microarray hybridization sample preparation
Abstract
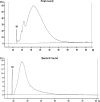
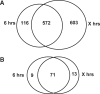
Gene expression analysis performed through comparative abundance of transcripts is facing a new challenge with the increasing need to compare samples of known cell number, such as early embryos or laser microbiopsies, where the RNA contents of identical cellular inputs can by nature be variable. When working with scarce tissues, the success of microarray profiling largely depends on the efficiency of the amplification step as determined by its ability to preserve the relative abundance of transcripts in the resulting amplified sample. Maintaining this initial relative abundance across samples is paramount to the generation of physiologically relevant data when comparing samples of different RNA content. The T7 RNA polymerase (T7-IVT) amplification is widely used for microarray sample preparation. Characterization of the reaction's kinetics has clearly indicated that its true linear phase is of short duration and is followed by a nonlinear phase. This second phase leads to modifications in transcript abundance that biases comparison between samples of different types. The impact assessment performed in this study has shown that the standard amplification protocol significantly lowers the quality of microarray data, rendering more than half of differentially expressed candidates undetected and distorting the true proportional differences of all candidates analyzed.
Figures





References
-
- Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. - PubMed
-
- Puskas LG, Zvara A, Hackler L, Jr, Van Hummelen P. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32:1330–1334. 1336, 1338, 1340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

