New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites
- PMID: 19336450
- PMCID: PMC2704433
- DOI: 10.1101/gr.089367.108
New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites
Abstract
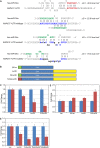
MicroRNAs (miRNAs) are known to post-transcriptionally regulate target mRNAs through the 3'-UTR, which interacts mainly with the 5'-end of miRNA in animals. Here we identify many endogenous motifs within human 5'-UTRs specific to the 3'-ends of miRNAs. The 3'-end of conserved miRNAs in particular has significant interaction sites in the human-enriched, less conserved 5'-UTR miRNA motifs, while human-specific miRNAs have significant interaction sites only in the conserved 5'-UTR motifs, implying both miRNA and 5'-UTR are actively evolving in response to each other. Additionally, many miRNAs with their 3'-end interaction sites in the 5'-UTRs turn out to simultaneously contain 5'-end interaction sites in the 3'-UTRs. Based on these findings we demonstrate combinatory interactions between a single miRNA and both end regions of an mRNA using model systems. We further show that genes exhibiting large-scale protein changes due to miRNA overexpression or deletion contain both UTR interaction sites predicted. We provide the predicted targets of this new miRNA target class, miBridge, as an efficient way to screen potential targets, especially for nonconserved miRNAs, since the target search space is reduced by an order of magnitude compared with the 3'-UTR alone. Efficacy is confirmed by showing SEC24D regulation with hsa-miR-605, a miRNA identified only in primate, opening the door to the study of nonconserved miRNAs. Finally, miRNAs (and associated proteins) involved in this new targeting class may prevent 40S ribosome scanning through the 5'-UTR and keep it from reaching the start-codon, preventing 60S association.
Figures




References
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. - PubMed
-
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
