Mutations in the beta-myosin rod cause myosin storage myopathy via multiple mechanisms
- PMID: 19336582
- PMCID: PMC2669361
- DOI: 10.1073/pnas.0900107106
Mutations in the beta-myosin rod cause myosin storage myopathy via multiple mechanisms
Abstract
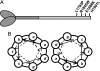
Myosin storage myopathy (MSM) is a congenital myopathy characterized by the presence of subsarcolemmal inclusions of myosin in the majority of type I muscle fibers, and has been linked to 4 mutations in the slow/cardiac muscle myosin, beta-MyHC (MYH7). Although the majority of the >230 disease causing mutations in MYH7 are located in the globular head region of the molecule, those responsible for MSM are part of a subset of MYH7 mutations that are located in the alpha-helical coiled-coil tail. Mutations in the myosin head are thought to affect the ATPase and actin-binding properties of the molecule. To date, however, there are no reports of the molecular mechanism of pathogenesis for mutations in the rod region of muscle myosins. Here, we present analysis of 4 mutations responsible for MSM: L1793P, R1845W, E1886K, and H1901L. We show that each MSM mutation has a different molecular phenotype, suggesting that there are multiple mechanisms by which MSM can be caused. These mechanisms range from thermodynamic and functional irregularities of individual proteins (L1793P), to varying defects in the assembly and stability of filaments formed from the proteins (R1845W, E1886K, and H1901L). In addition to furthering our understanding of MSM, these observations provide the first insight into how mutations affect the rod region of muscle myosins, and provide a framework for future studies of disease-causing mutations in this region of the molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
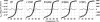
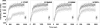
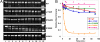

References
-
- Atkinson SJ, Stewart M. Molecular interactions in myosin assembly. Role of the 28-residue charge repeat in the rod. J Mol Biol. 1992;226:7–13. - PubMed
-
- Atkinson SJ, Stewart M. Molecular basis of myosin assembly: Coiled-coil interactions and the role of charge periodicities. J Cell Sci Suppl. 1991;14:7–10. - PubMed
-
- McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. - PubMed
-
- Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

