Codanin-1, the protein encoded by the gene mutated in congenital dyserythropoietic anemia type I (CDAN1), is cell cycle-regulated
- PMID: 19336738
- PMCID: PMC2675674
- DOI: 10.3324/haematol.2008.003327
Codanin-1, the protein encoded by the gene mutated in congenital dyserythropoietic anemia type I (CDAN1), is cell cycle-regulated
Abstract
Background: Congenital dyserythropoietic anemia type I is an inherited autosomal recessive macrocytic anemia associated with ineffective erythropoiesis and the development of secondary hemochromatosis. Distinct erythroid precursors with internuclear chromatin bridges and spongy heterochromatin are pathognomonic for the disease. The mutated gene (CDAN1) encodes a ubiquitously expressed protein of unknown function, codanin-1. Based on the morphological features of congenital dyserythropoietic anemia type I erythroblasts and data on a role in cell cycle progression of codanin-1 homolog in Drosophila we investigated the cellular localization and possible involvement of codanin-1 during the cell cycle.
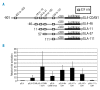
Design and methods: Codanin-1 localization was studied by immunofluorescence and immune electron microscopy. Cell cycle expression of codanin-1 was evaluated using synchronized HeLa cells. E2F proteins are the main regulator of G(1)/S transition. An E2F1-inducible cell line (U20S-ER-E2F1) enabled us to study codanin-1 expression following ectopic E2F1 induction. Direct binding of E2F1 to codanin-1 promoter was assessed by chromatin immunoprecipitation. We used a luciferase-reporter plasmid to study activation of CDAN1 transcription by E2F1.
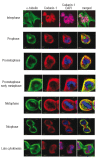
Results: We localized codanin-1 to heterochromatin in interphase cells. During the cell cycle, high levels of codanin-1 were observed in the S phase. At mitosis, codanin-1 underwent phosphorylation, which coincided with its exclusion from condensed chromosomes. The proximal CDAN1 gene promoter region, containing five putative E2F binding sites, was found to be a direct target of E2F1.
Conclusions: Taken together, these data suggest that codanin-1 is a cell cycle-regulated protein active in the S phase. The exact role of codanin-1 during the S phase remains to be determined. Nevertheless this represents the first step towards understanding the function of the proteins involved in congenital dyserythropoietic anemia.
Figures




Comment in
-
Close to unraveling the secrets of congenital dyserythropoietic anemia types I and II.Haematologica. 2009 May;94(5):599-602. doi: 10.3324/haematol.2009.005785. Haematologica. 2009. PMID: 19407313 Free PMC article.
Similar articles
-
Clinical aspects and pathogenesis of congenital dyserythropoietic anemias: from morphology to molecular approach.Haematologica. 2012 Dec;97(12):1786-94. doi: 10.3324/haematol.2012.072207. Epub 2012 Oct 12. Haematologica. 2012. PMID: 23065504 Free PMC article. Review.
-
Characterization of the interactions between Codanin-1 and C15Orf41, two proteins implicated in congenital dyserythropoietic anemia type I disease.BMC Mol Cell Biol. 2020 Mar 23;21(1):18. doi: 10.1186/s12860-020-00258-1. BMC Mol Cell Biol. 2020. PMID: 32293259 Free PMC article.
-
Congenital dyserythropoietic anemia type I is caused by mutations in codanin-1.Am J Hum Genet. 2002 Dec;71(6):1467-74. doi: 10.1086/344781. Epub 2002 Nov 14. Am J Hum Genet. 2002. PMID: 12434312 Free PMC article.
-
Codanin-1 mutations in congenital dyserythropoietic anemia type 1 affect HP1{alpha} localization in erythroblasts.Blood. 2011 Jun 23;117(25):6928-38. doi: 10.1182/blood-2010-09-308478. Epub 2011 Mar 1. Blood. 2011. PMID: 21364188
-
Congenital dyserythropoietic anemia in China: a case report from two families and a review.Ann Hematol. 2014 May;93(5):773-7. doi: 10.1007/s00277-013-1933-8. Epub 2013 Nov 7. Ann Hematol. 2014. PMID: 24196372 Review.
Cited by
-
Clinical aspects and pathogenesis of congenital dyserythropoietic anemias: from morphology to molecular approach.Haematologica. 2012 Dec;97(12):1786-94. doi: 10.3324/haematol.2012.072207. Epub 2012 Oct 12. Haematologica. 2012. PMID: 23065504 Free PMC article. Review.
-
Congenital dyserythropoietic anemia.Int J Hematol. 2010 Oct;92(3):432-8. doi: 10.1007/s12185-010-0667-9. Epub 2010 Sep 7. Int J Hematol. 2010. PMID: 20820969
-
Congenital dyserythropoietic anemias: molecular insights and diagnostic approach.Blood. 2013 Sep 26;122(13):2162-6. doi: 10.1182/blood-2013-05-468223. Epub 2013 Aug 12. Blood. 2013. PMID: 23940284 Free PMC article. Review.
-
Mild dyserythropoiesis and β-like globin gene expression imbalance due to the loss of histone chaperone ASF1B.Hum Genomics. 2020 Oct 16;14(1):39. doi: 10.1186/s40246-020-00283-3. Hum Genomics. 2020. PMID: 33066815 Free PMC article.
-
Close to unraveling the secrets of congenital dyserythropoietic anemia types I and II.Haematologica. 2009 May;94(5):599-602. doi: 10.3324/haematol.2009.005785. Haematologica. 2009. PMID: 19407313 Free PMC article.
References
-
- Heimpel H, Wendt F. Congenital dyserythropoietic anemia with karyorrhexis and multinuclearity of erythroblasts. Helv Med Acta. 1968;34:103–15. - PubMed
-
- Sandstrom H, Wahlin A. Congenital dyserythropoietic anemia type III. Haematologica. 2000;85:753–7. - PubMed
-
- Lind L, Sandstrom H, Wahlin A, Eriksson M, Nilsson-Sojka B, Sikstrom C, et al. Localization of the gene for congenital dyserythropoietic anemia type III, CDAN3, to chromosome 15q21-q25. Hum Mol Genet. 1995;4:109–12. - PubMed
-
- Delaunay J, Iolascon A. The congenital dyserythropoietic anaemias. Baillieres Best Pract Res Clin Haematol. 1999;12:691–705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

