Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements
- PMID: 19337366
- PMCID: PMC2659449
- DOI: 10.1371/journal.pone.0005103
Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements
Abstract
Background: More than 80% of mammalian protein-coding genes are driven by TATA-less promoters which often show multiple transcriptional start sites (TSSs). However, little is known about the core promoter DNA sequences or mechanisms of transcriptional initiation for this class of promoters.
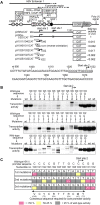
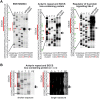
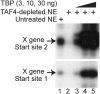
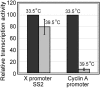
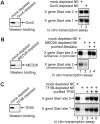
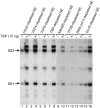
Methodology/principal findings: Here we identify a new core promoter element XCPE2 (X core promoter element 2) (consensus sequence: A/C/G-C-C/T-C-G/A-T-T-G/A-C-C/A(+1)-C/T) that can direct specific transcription from the second TSS of hepatitis B virus X gene mRNA. XCPE2 sequences can also be found in human promoter regions and typically appear to drive one of the start sites within multiple TSS-containing TATA-less promoters. To gain insight into mechanisms of transcriptional initiation from this class of promoters, we examined requirements of several general transcription factors by in vitro transcription experiments using immunodepleted nuclear extracts and purified factors. Our results show that XCPE2-driven transcription uses at least TFIIB, either TFIID or free TBP, RNA polymerase II (RNA pol II) and the MED26-containing mediator complex but not Gcn5. Therefore, XCPE2-driven transcription can be carried out by a mechanism which differs from previously described TAF-dependent mechanisms for initiator (Inr)- or downstream promoter element (DPE)-containing promoters, the TBP- and SAGA (Spt-Ada-Gcn5-acetyltransferase)-dependent mechanism for yeast TATA-containing promoters, or the TFTC (TBP-free-TAF-containing complex)-dependent mechanism for certain Inr-containing TATA-less promoters. EMSA assays using XCPE2 promoter and purified factors further suggest that XCPE2 promoter recognition requires a set of factors different from those for TATA box, Inr, or DPE promoter recognition.
Conclusions/significance: We identified a new core promoter element XCPE2 that are found in multiple TSS-containing TATA-less promoters. Mechanisms of promoter recognition and transcriptional initiation for XCPE2-driven promoters appear different from previously shown mechanisms for classical promoters that show single "focused" TSSs. Our studies provide insight into novel mechanisms of RNA Pol II transcription from multiple TSS-containing TATA-less promoters.
Conflict of interest statement
Figures




References
-
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. - PubMed
-
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

