Exploring writhe in supercoiled minicircle DNA
- PMID: 19337583
- PMCID: PMC2662687
- DOI: 10.1088/0953-8984/18/14/S01
Exploring writhe in supercoiled minicircle DNA
Abstract
Using λ-Int recombination in E. coli, we have generated milligram quantities of supercoiled minicircle DNA. Intramolecular Int recombination was efficient down to lengths ~254 bp. When nicked and religated in the presence of ethidium bromide, 339 bp minicircles adopted at least seven unique topoisomers that presumably correspond to ΔLk ranging from 0 to -6, which we purified individually. We used these minicircles, with unique ΔLk, to address the partition into twist and writhe as a function of ΔLk. Gel electrophoresis and atomic force microscopy revealed progressively higher writhe conformations in the presence of 10 mM CaCl(2) or MgCl(2). From simplistic calculations of the bending and twisting energies, we predict the elastic free energy of supercoiling for these minicircles to be lower than if the supercoiling was partitioned mainly into twist. The predicted writhe corresponds closely with that which we observed experimentally in the presence of divalent metal ions. However, in the absence of divalent metal ions only limited writhe was observed, demonstrating the importance of electrostatic effects on DNA structure, when the screening of charges on the DNA is weak. This study represents a unique insight into the supercoiling of minicircle DNA, with implications for DNA structure in general.
Figures


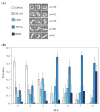
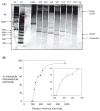

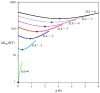
References
-
- Bates AD, Maxwell A. DNA Topology. 2. Oxford: Oxford University Press; 2005.
-
- Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD. J Mol Biol. 1994;235:825–47. - PubMed
-
- Bliska JB, Cozzarelli NR. J Mol Biol. 1987;194:205–18. - PubMed
-
- Cloutier TE, Widom J. Mol Cell. 2004;14:355–62. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
