Snapshot peptidomics of the regulated secretory pathway
- PMID: 19339239
- PMCID: PMC2709191
- DOI: 10.1074/mcp.M900044-MCP200
Snapshot peptidomics of the regulated secretory pathway
Abstract
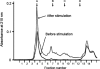
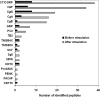
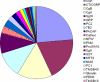
Neurons and endocrine cells have the regulated secretory pathway (RSP) in which precursor proteins undergo proteolytic processing by prohormone convertase (PC) 1/3 or 2 to generate bioactive peptides. Although motifs for PC-mediated processing have been described ((R/K)X(n)(R/K) where n = 0, 2, 4, or 6), actual processing sites cannot be predicted from amino acid sequences alone. We hypothesized that discovery of bioactive peptides would be facilitated by experimentally identifying signal peptide cleavage sites and processing sites. However, in vivo and in vitro peptide degradation, which is widely recognized in peptidomics, often hampers processing site determination. To obtain sequence information about peptides generated in the RSP on a large scale, we applied a brief exocytotic stimulus (2 min) to cultured endocrine cells and analyzed peptides released into supernatant using LC-MSMS. Of note, 387 of the 400 identified peptides arose from 19 precursor proteins known to be processed in the RSP, including nine peptide hormone and neuropeptide precursors, seven granin-like proteins, and three processing enzymes (PC1/3, PC2, and peptidyl-glycine alpha-amidating monooxygenase). In total, 373 peptides were informative enough to predict processing sites in that they have signal sequence cleavage sites, PC consensus sites, or monobasic cleavage sites. Several monobasic cleavage sites identified here were previously proved to be generated by PCs. Thus, our approach helps to predict processing sites of RSP precursor proteins and will expedite the identification of unknown bioactive peptides hidden in precursor sequences.
Figures

References
-
- Zhou A., Webb G., Zhu X., Steiner D. F. ( 1999) Proteolytic processing in the secretory pathway. J. Biol. Chem. 274, 20745– 20748 - PubMed
-
- Brakch N., Rholam M., Boussetta H., Cohen P. ( 1993) Role of beta-turn in proteolytic processing of peptide hormone precursors at dibasic sites. Biochemistry 32, 4925– 4930 - PubMed
-
- Clynen E., Baggerman G., Veelaert D., Cerstiaens A., Van der Horst D., Harthoorn L., Derua R., Waelkens E., De Loof A., Schoofs L. ( 2001) Peptidomics of the pars intercerebralis-corpus cardiacum complex of the migratory locust, Locusta migratoria. Eur. J. Biochem. 268, 1929– 1939 - PubMed
-
- Schrader M., Schulz-Knappe P. ( 2001) Peptidomics technologies for human body fluids. Trends Biotechnol. 19, S 55– 60 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

