Potential role of phenotypic mutations in the evolution of protein expression and stability
- PMID: 19339491
- PMCID: PMC2669386
- DOI: 10.1073/pnas.0809506106
Potential role of phenotypic mutations in the evolution of protein expression and stability
Abstract
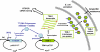
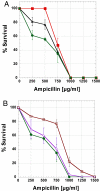
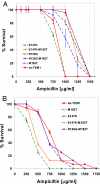
Phenotypic mutations (errors occurring during protein synthesis) are orders of magnitude more frequent than genetic mutations. Consequently, the sequences of individual protein molecules transcribed and translated from the same gene can differ. To test the effects of such mutations, we established a bacterial system in which an antibiotic resistance gene (TEM-1 beta-lactamase) was transcribed by either a high-fidelity RNA polymerase or its error-prone mutant. This setup enabled the analysis of individual mRNA transcripts that were synthesized under normal or error-prone conditions. We found that an increase of approximately 20-fold in the frequency of transcription errors promoted the evolution of higher TEM-1 expression levels and of more stable enzyme variants. The stabilized variants exhibited a distinct advantage under error-prone transcription, although under normal transcription they conferred resistance similar to wild-type TEM-1. They did so, primarily, by increasing TEM-1's tolerance to destabilizing deleterious mutations that arise from transcriptional errors. The stabilized TEM-1 variants also showed increased tolerance to genetic mutations. Thus, although phenotypic mutations are not individually subjected to inheritance and natural selection, as are genetic mutations, they collectively exert a direct and immediate effect on protein fitness. They may therefore play a role in shaping protein traits such as expression levels, stability, and tolerance to genetic mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
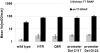
References
-
- Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. - PubMed
-
- Ellis N, Gallant J. An estimate of the global error frequency in translation. Mol Gen Genet. 1982;188:169–172. - PubMed
-
- Rosenberger RF, Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191:207–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

