Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop
- PMID: 19339507
- PMCID: PMC2689985
- DOI: 10.1104/pp.108.132787
Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop
Abstract
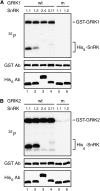
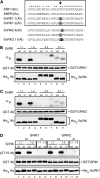
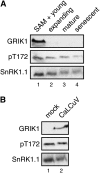
SNF1-related kinases (SnRK1s) play central roles in coordinating energy balance and nutrient metabolism in plants. SNF1 and AMPK, the SnRK1 homologs in budding yeast (Saccharomyces cerevisiae) and mammals, are activated by phosphorylation of conserved threonine residues in their activation loops. Arabidopsis (Arabidopsis thaliana) GRIK1 and GRIK2, which were first characterized as geminivirus Rep interacting kinases, are phylogenetically related to SNF1 and AMPK activating kinases. In this study, we used recombinant proteins produced in bacteria to show that both GRIKs specifically bind to the SnRK1 catalytic subunit and phosphorylate the equivalent threonine residue in its activation loop in vitro. GRIK-mediated phosphorylation increased SnRK1 kinase activity in autophosphorylation and peptide substrate assays. These data, together with earlier observations that GRIKs could complement yeast mutants lacking SNF1 activation activities, established that the GRIKs are SnRK1 activating kinases. Given that the GRIK proteins only accumulate in young tissues and geminivirus-infected mature leaves, the GRIK-SnRK1 cascade may function in a developmentally regulated fashion and coordinate the unique metabolic requirements of rapidly growing cells and geminivirus-infected cells that have been induced to reenter the cell cycle.
Figures



Similar articles
-
Upstream kinases of plant SnRKs are involved in salt stress tolerance.Plant J. 2018 Jan;93(1):107-118. doi: 10.1111/tpj.13761. Epub 2017 Dec 2. Plant J. 2018. PMID: 29094495 Free PMC article.
-
Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases.Plant Physiol. 2006 Dec;142(4):1642-55. doi: 10.1104/pp.106.088476. Epub 2006 Oct 13. Plant Physiol. 2006. PMID: 17041027 Free PMC article.
-
Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5'-AMP.Plant J. 1999 Aug;19(4):433-9. doi: 10.1046/j.1365-313x.1999.00532.x. Plant J. 1999. PMID: 10504565
-
Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1.Trends Plant Sci. 2016 Apr;21(4):341-353. doi: 10.1016/j.tplants.2015.11.001. Epub 2015 Nov 28. Trends Plant Sci. 2016. PMID: 26642889 Review.
-
Plant SnRK1 Kinases: Structure, Regulation, and Function.Exp Suppl. 2016;107:403-438. doi: 10.1007/978-3-319-43589-3_17. Exp Suppl. 2016. PMID: 27812990 Review.
Cited by
-
Begomoviruses associated with okra yellow vein mosaic disease (OYVMD): diversity, transmission mechanism, and management strategies.Mol Hortic. 2024 Nov 5;4(1):36. doi: 10.1186/s43897-024-00112-4. Mol Hortic. 2024. PMID: 39497157 Free PMC article. Review.
-
Small RNAs from the plant pathogenic fungus Sclerotinia sclerotiorum highlight host candidate genes associated with quantitative disease resistance.Mol Plant Pathol. 2019 Sep;20(9):1279-1297. doi: 10.1111/mpp.12841. Epub 2019 Jul 30. Mol Plant Pathol. 2019. PMID: 31361080 Free PMC article.
-
Arabidopsis CTP:phosphocholine cytidylyltransferase 1 is phosphorylated and inhibited by sucrose nonfermenting 1-related protein kinase 1 (SnRK1).J Biol Chem. 2019 Oct 25;294(43):15862-15874. doi: 10.1074/jbc.RA119.008047. Epub 2019 Aug 22. J Biol Chem. 2019. PMID: 31439667 Free PMC article.
-
Quantitative Proteomic Analysis of the Response to Cold Stress in Jojoba, a Tropical Woody Crop.Int J Mol Sci. 2019 Jan 9;20(2):243. doi: 10.3390/ijms20020243. Int J Mol Sci. 2019. PMID: 30634475 Free PMC article.
-
A plant kinase plays roles in defense response against geminivirus by phosphorylation of a viral pathogenesis protein.Plant Signal Behav. 2012 Jul;7(7):888-92. doi: 10.4161/psb.20646. Epub 2012 Jul 1. Plant Signal Behav. 2012. PMID: 22751295 Free PMC article.
References
-
- Alessi DR, Sakamoto K, Bayascas JR (2006) LKB1-dependent signaling pathways. Annu Rev Biochem 75 137–163 - PubMed
-
- Ascencio-Ibañez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting the pathogen response and cell cycle controls during geminivirus infection. Plant Physiol 148 436–454 - PMC - PubMed
-
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

