Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization
- PMID: 19339989
- PMCID: PMC2688536
- DOI: 10.1038/emboj.2009.86
Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization
Abstract
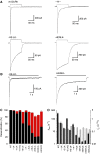
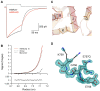
AMPA and kainate receptors mediate fast synaptic transmission. AMPA receptor ligand-binding domains form dimers, which are key functional units controlling ion-channel activation and desensitization. Dimer stability is inversely related to the rate and extent of desensitization. Kainate and AMPA receptors share common structural elements, but functional measurements suggest that subunit assembly and gating differs between these subtypes. To investigate this, we constructed a library of GluR6 kainate receptor mutants and directly measured changes in kainate receptor dimer stability by analytical ultracentrifugation, which, combined with electrophysiological experiments, revealed an inverse correlation between dimer stability and the rate of desensitization. We solved crystal structures for a series of five GluR6 mutants, to understand the molecular mechanisms for dimer stabilization. We demonstrate that the desensitized state of kainate receptors acts as a deep energy well offsetting the stabilizing effects of dimer interface mutants, and that the deactivation of kainate receptor responses is dominated by entry into desensitized states. Our results show how neurotransmitter receptors with similar structures and gating mechanisms can exhibit strikingly different functional properties.
Figures







References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Armstrong N, Gouaux E (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28: 165–181 - PubMed
-
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127: 85–97 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

