Targeting and function of the mitochondrial fission factor GDAP1 are dependent on its tail-anchor
- PMID: 19340293
- PMCID: PMC2659752
- DOI: 10.1371/journal.pone.0005160
Targeting and function of the mitochondrial fission factor GDAP1 are dependent on its tail-anchor
Abstract
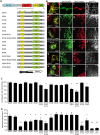
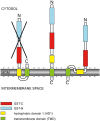
Proteins controlling mitochondrial dynamics are often targeted to and anchored into the mitochondrial outer membrane (MOM) by their carboxyl-terminal tail-anchor domain (TA). However, it is not known whether the TA modulates protein function. GDAP1 is a mitochondrial fission factor with two neighboring hydrophobic domains each flanked by basic amino acids (aa). Here we define GDAP1 as TA MOM protein. GDAP1 carries a single transmembrane domain (TMD) that is, together with the adjacent basic aa, critical for MOM targeting. The flanking N-terminal region containing the other hydrophobic domain is located in the cytoplasm. TMD sequence, length, and high hydrophobicity do not influence GDAP1 fission function if MOM targeting is maintained. The basic aa bordering the TMD in the cytoplasm, however, are required for both targeting of GDAP1 as part of the TA and GDAP1-mediated fission. Thus, this GDAP1 region contains critical overlapping motifs defining intracellular targeting by the TA concomitant with functional aspects.
Conflict of interest statement
Figures





References
-
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. - PubMed
-
- Niemann A, Berger P, Suter U. Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:217–242. - PubMed
-
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, et al. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. - PubMed
-
- Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

