Integrated capture, concentration, polymerase chain reaction, and capillary electrophoretic analysis of pathogens on a chip
- PMID: 19341275
- PMCID: PMC2684796
- DOI: 10.1021/ac900060r
Integrated capture, concentration, polymerase chain reaction, and capillary electrophoretic analysis of pathogens on a chip
Abstract
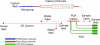
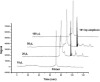
A laboratory-on-a-chip system for pathogen detection is presented that integrates cell preconcentration, purification, polymerase chain reaction (PCR), and capillary electrophoretic (CE) analysis. The microdevice is composed of micropumps and valves, a cell capture structure, a 100 nL PCR reactor, and a 5 cm long CE column for amplicon separation. Sample volumes ranging from 10 to 100 microL are introduced and driven through a fluidized bed of magnetically constrained immunomagnetic beads where the target cells are captured. After cell capture, beads are transferred using the on-chip pumps to the PCR reactor for DNA amplification. The resulting PCR products are electrophoretically injected onto the CE column for separation and detection of Escherichia coli K12 and E. coli O157 targets. A detection limit of 0.2 cfu/microL is achieved using the E. coli O157 target and an input volume of 50 microL. Finally, the sensitive detection of E. coli O157 in the presence of K12 at a ratio of 1:1000 illustrates the capability of our system to identify target cells in a high commensal background. This cell capture-PCR-CE microsystem is a significant advance in the development of rapid, sensitive, and specific laboratory-on-a-chip devices for pathogen detection.
Figures



References
-
- Lagally ET, Mathies RA. J. Phys. D: Appl. Phys. 2004;37:245–261.
-
- Liu P, Seo TS, Beyor N, Shin KJ, Scherer JR, Mathies RA. Anal. Chem. 2007;79:1881–1889. - PubMed
-
- Seiler K, Harrison DJ, Manz A. Anal. Chem. 1993;65:1481–1488.
-
- Ocvirk G, Tang T, Harrison DJ. Analyst. Vol. 123. Cambridge, U. K.: 1998. pp. 1429–1434.
-
- Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Anal. Chem. 2007;79:8557–8563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

