MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus
- PMID: 19342367
- PMCID: PMC2671281
- DOI: 10.2353/ajpath.2009.080718
MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus
Abstract
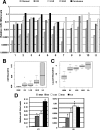
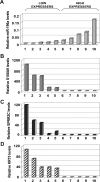
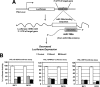
Barrett's esophagus (BE)/Barrett's metaplasia (BM) is a recognized precursor of esophageal adenocarcinoma (EA) with an intermediary stage of dysplasia. The low yield and high cost of endoscopic screening of patients with BE underscores the need for novel biomarkers, such as microRNA (miRNA), which have emerged as important players in neoplastic progression for risk assessment of developing dysplasia/adenocarcinoma. Recently, we reported highly elevated levels of miRNA-196a (miR-196a) in EA and demonstrated its growth-promoting and anti-apoptotic functions. Here, we evaluated miR-196a as a marker of BE progression to low-grade dysplasia, high-grade dysplasia, and EA using microdissected paraffin-embedded tissues from 11 patients. Higher levels of miR-196a were observed in EA, BE, and dysplastic lesions compared with normal squamous mucosa, and in high-grade dysplasia compared with BE and low-grade dysplasia. Using frozen tumor tissues from 10 additional patients who had advanced EA, we evaluated the correlation of miR-196a with its in silico-predicted targets, keratin 5 (KRT5), small proline-rich protein 2C (SPRR2C), and S100 calcium-binding protein A9 (S100A9), which are down-regulated during BE progression. MiR-196a levels inversely correlated with the predicted target mRNA levels in EA. We confirmed that miR-196a specifically targets KRT5, SPRR2C, and S100A9 3' UTRs using miR-196a-mimic and luciferase reporter-based assays. In conclusion, this study identified miR-196a as a potential marker of progression of BE and KRT5, SPRR2C, and S100A9 as its targets.
Figures


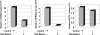
References
-
- Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. - PubMed
-
- Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in Barrett’s esophagus? Gastroenterology. 2000;119:333–338. - PubMed
-
- Swisher SG, Hunt KK, Holmes EC, Zinner MJ, McFadden DW. Changes in the surgical management of esophageal cancer from 1970 to 1993. Am J Surg. 1995;169:609–614. - PubMed
-
- Reid BJ, Prevo LJ, Galipeau PC, Sanchez CA, Longton G, Levine DS, Blount PL, Rabinovitch PS. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. - PMC - PubMed
-
- Reid BJ, Blount PL, Rabinovitch PS. Biomarkers in Barrett’s esophagus. Gastrointest Endosc Clin N Am. 2003;13:369–397. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

