Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway
- PMID: 19342893
- PMCID: PMC2865178
- DOI: 10.4161/cc.8.9.8215
Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway
Abstract
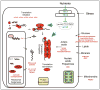
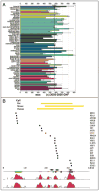
Among other signals, cell growth is particularly controlled by the target of rapamycin (TOR) pathway that includes the tuberous sclerosis complex genes (TSC1/2), and through transcriptional effects regulated by c-myc. Overexpression of Drosophila Myc and TSC1/2 cause opposing growth and proliferation defects. Despite this relationship, direct regulatory connections between Myc and the TSC have only recently been evaluated. Other than studies of p53 regulation, little consideration has been given to transcriptional regulation of the TSC genes. Here we review evidence that transcriptional controls are potentially important regulators of TSC2 expression, and that Myc is a direct repressor of its expression. Since tuberin loss de-represses Myc protein, the connection between these two growth regulators is positioned to act as a feed-forward loop that would amplify the oncogenic effects of decreased tuberin or increased Myc. Further experiments will be needed to clarify the mechanisms underlying this important connection, and evaluate its overall contribution to cancers caused by TSC loss or Myc gain.
Figures


Similar articles
-
Mammalian target of rapamycin complex 1 (mTORC1) enhances bortezomib-induced death in tuberous sclerosis complex (TSC)-null cells by a c-MYC-dependent induction of the unfolded protein response.J Biol Chem. 2013 May 31;288(22):15687-98. doi: 10.1074/jbc.M112.431056. Epub 2013 Apr 23. J Biol Chem. 2013. PMID: 23612979 Free PMC article.
-
Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb.Curr Biol. 2003 Aug 5;13(15):1259-68. doi: 10.1016/s0960-9822(03)00506-2. Curr Biol. 2003. PMID: 12906785
-
c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation.Cancer Res. 2007 Dec 1;67(23):11209-17. doi: 10.1158/0008-5472.CAN-06-4351. Cancer Res. 2007. PMID: 18056446 Free PMC article.
-
Tumour suppressors hamartin and tuberin: intracellular signalling.Cell Signal. 2003 Aug;15(8):729-39. doi: 10.1016/s0898-6568(03)00040-8. Cell Signal. 2003. PMID: 12781866 Review.
-
Tuberous sclerosis: from tubers to mTOR.Ann Hum Genet. 2003 Jan;67(Pt 1):87-96. doi: 10.1046/j.1469-1809.2003.00012.x. Ann Hum Genet. 2003. PMID: 12556239 Review.
Cited by
-
An IRF4-MYC-mTORC1 Integrated Pathway Controls Cell Growth and the Proliferative Capacity of Activated B Cells during B Cell Differentiation In Vivo.J Immunol. 2021 Oct 1;207(7):1798-1811. doi: 10.4049/jimmunol.2100440. Epub 2021 Sep 1. J Immunol. 2021. PMID: 34470852 Free PMC article.
-
Regulation of gene expression in hepatic cells by the mammalian Target of Rapamycin (mTOR).PLoS One. 2010 Feb 5;5(2):e9084. doi: 10.1371/journal.pone.0009084. PLoS One. 2010. PMID: 20140209 Free PMC article.
-
Ribosomal Protein S6: A Potential Therapeutic Target against Cancer?Int J Mol Sci. 2021 Dec 21;23(1):48. doi: 10.3390/ijms23010048. Int J Mol Sci. 2021. PMID: 35008473 Free PMC article. Review.
-
Upregulation of the pathogenic transcription factor SPI1/PU.1 in tuberous sclerosis complex and focal cortical dysplasia by oxidative stress.Brain Pathol. 2021 Sep;31(5):e12949. doi: 10.1111/bpa.12949. Epub 2021 Mar 30. Brain Pathol. 2021. PMID: 33786950 Free PMC article.
-
p62/SQSTM1 enhances breast cancer stem-like properties by stabilizing MYC mRNA.Oncogene. 2017 Jan 19;36(3):304-317. doi: 10.1038/onc.2016.202. Epub 2016 Jun 27. Oncogene. 2017. PMID: 27345399 Free PMC article.
References
-
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. - PubMed
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Mitchison JM. The biology of the cell cycle. Cambridge [Eng.]: University Press; 1971.
-
- Hartwell LH. Genetic control of the cell division cycle in yeast II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
