Structure-function relationships in peptoids: recent advances toward deciphering the structural requirements for biological function
- PMID: 19343235
- PMCID: PMC5962266
- DOI: 10.1039/b817980h
Structure-function relationships in peptoids: recent advances toward deciphering the structural requirements for biological function
Abstract
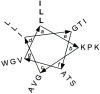
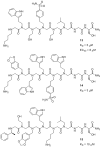
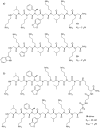
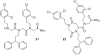
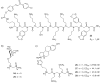
Oligomers of N-substituted glycine, or peptoids, are versatile tools to probe biological processes and hold promise as therapeutic agents. An underlying theme in the majority of recent peptoid research is the connection between peptoid function and peptoid structure. For certain applications, well-folded peptoids are essential for activity, while unstructured peptoids appear to suffice, or even are superior, for other applications. Currently, these structure-function connections are largely made after the design, synthesis, and characterization process. However, as guidelines for peptoid folding are elucidated and the known biological activities are expanded, we anticipate these connections will provide a pathway toward the de novo design of functional peptoids. In this perspective, we review several of the peptoid structure-function relationships that have been delineated over the past five years.
Figures
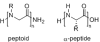
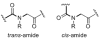
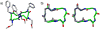
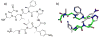
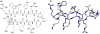
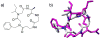
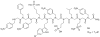
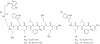
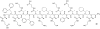
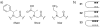

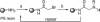
References
-
- Farmer PS, Ariëns EJ. Trends Pharmacol Sci. 1982;3:362–365.
-
- This concept was applied to the development of novel cholecystokinin (CCK) analogs by Horwell et al. Horwell DC, Beeby A, Clark CR, Hughes J, editors. J Med Chem. 1987;30:729. - PubMed
-
- Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J Am Chem Soc. 1992;114:10646–10647.
-
- Zuckermann RN, Martin EJ, Spellmeyer DC, Stauber GB, Shoemaker KR, Kerr JM, Figliozzi GM, Goff DA, Siani MA, Simon R, Banville SC, Brown EG, Wang L, Richter LS, Moos WH. J Med Chem. 1994;37:2678–2685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

