Noncytotoxic orange and red/green derivatives of DsRed-Express2 for whole-cell labeling
- PMID: 19344508
- PMCID: PMC2678115
- DOI: 10.1186/1472-6750-9-32
Noncytotoxic orange and red/green derivatives of DsRed-Express2 for whole-cell labeling
Abstract
Background: Whole-cell labeling is a common application of fluorescent proteins (FPs), but many red and orange FPs exhibit cytotoxicity that limits their use as whole-cell labels. Recently, a tetrameric red FP called DsRed-Express2 was engineered for enhanced solubility and was shown to be noncytotoxic in bacterial and mammalian cells. Our goal was to create derivatives of this protein with different spectral properties.
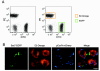
Results: Building on previous studies of DsRed mutants, we created two DsRed-Express2 derivatives: E2-Orange, an orange FP, and E2-Red/Green, a dual-color FP with both red and green emission. We show that these new FPs retain the low cytotoxicity of DsRed-Express2. In addition, we show that these new FPs are useful as second or third colors for flow cytometry and fluorescence microscopy.
Conclusion: E2-Orange and E2-Red/Green will facilitate the production of healthy, stably fluorescent cell lines and transgenic organisms for multi-color labeling studies.
Figures




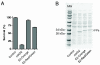

References
-
- Tao W, Evans BG, Yao J, Cooper S, Cornetta K, Ballas CB, Hangoc G, Broxmeyer HE. Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-Express fluorescent protein is not. Stem Cells. 2007;25:670–678. doi: 10.1634/stemcells.2006-0553. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

