Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function
- PMID: 19344517
- PMCID: PMC2678116
- DOI: 10.1186/1472-6750-9-33
Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function
Abstract
Background: Transgenic chloroplasts are potential bioreactors for recombinant protein production, especially for achievement of high levels of protein expression and proper folding. Production of therapeutic proteins in leaves provides transgene containment by elimination of reproductive structures. Therefore, in this study, human Insulin like Growth Factor-1 is expressed in transgenic chloroplasts for evaluation of structural identity and function.
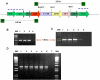
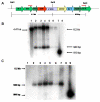
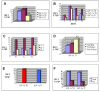
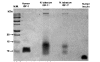
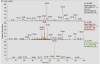
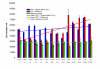
Results: Expression of the synthetic Insulin like Growth Factor 1 gene (IGF-1s, 60% AT) was observed in transformed E. coli. However, no native IGF-1 gene (IGF-1n, 41% AT) product was detected in the western blots in E. coli. Site-specific integration of the transgenes into the tobacco chloroplast genome was confirmed after transformation using PCR. Southern blot analysis confirmed that the transgenic lines were homoplasmic. The transgenic plant lines had IGF-1s expression levels of 11.3% of total soluble protein (TSP). The IGF-1n plants contained 9.5% TSP as IGF-1n, suggesting that the chloroplast translation machinery is more flexible than E. coli in codon preference and usage. The expression of IGF-1 was increased up to 32% TSP under continuous illumination by the chloroplast light regulatory elements. IgG-Sepharose affinity column chromatographic separation of Z domain containing chloroplast derived IGF-1 protein, single and two dimensional electrophoresis methods and mass spectrometer analysis confirmed the identity of human IGF-1 in transgenic chloroplasts. Two spots analyzed from 2-D focusing/phoresis acrylamide gel showed the correct amino acid sequence of human IGF-1 and the S. aureus Z-tag. Cell proliferation assays in human HU-3 cells demonstrated the biological activity of chloroplast derived IGF-1 even in the presence of the S. aureus Z tag.
Conclusion: This study demonstrates that the human Insulin like Growth Factor-1 expressed in transgenic chloroplasts is identical to the native protein and is fully functional. The ability to use plant chloroplasts as bioreactors to generate proteins of great economic value that retain their biological activity is an exciting and achievable goal that appears to be within our grasp.
Figures






References
-
- Jones JH, Clemmons DR. Insulin like growth factors and their binding proteins: biological functions. Endocr Rev. 1995;16:3–10. - PubMed
-
- Feld S, Hirschberg R. Growth hormone, the insulin-like growth factor system, and the kidney. Endocr Rev. 1996;17:423–480. - PubMed
-
- Walsh G. Biopharmaceuticals: biochemistry and biotechnology. 1. John Wiley and Sons, England; 1998. pp. 235–243.
-
- Kim S, Lee Y. High-level expression and simple purification of recombinant human insulin-like growth factor I. J Biotechnol. 1996;48:97–105. - PubMed
-
- Blomsma MC, de Knegt RJ, Dullaart RP, Jansen PL. Insulin-like growth factor- I in liver cirrhosis. J Hepatology. 1997;27:1133–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

