Replication timing and transcriptional control: beyond cause and effect--part II
- PMID: 19345088
- PMCID: PMC2677117
- DOI: 10.1016/j.gde.2009.02.002
Replication timing and transcriptional control: beyond cause and effect--part II
Abstract
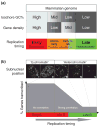
Replication timing is frequently discussed superficially in terms of its relationship to transcriptional activity via chromatin structure. However, so little is known about what regulates where and when replication initiates that it has been impossible to identify mechanistic and causal relationships. Moreover, much of our knowledge base has been anecdotal, derived from analyses of a few genes in unrelated cell lines. Recent studies have revisited long-standing hypotheses using genome-wide approaches. In particular, the foundation of this field was recently shored up with incontrovertible evidence that cellular differentiation is accompanied by coordinated changes in replication timing and transcription. These changes accompany subnuclear repositioning, and take place at the level of megabase-sized domains that transcend localized changes in chromatin structure or transcription. Inferring from these results, we propose that there exists a key transition during the middle of S-phase and that changes in replication timing traversing this period are associated with subnuclear repositioning and changes in the activity of certain classes of promoters.
Figures
References
-
- Schubeler D, Scalzo D, Kooperberg C, Van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32:438–442. - PubMed
-
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

