In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy
- PMID: 19345105
- PMCID: PMC2832475
- DOI: 10.1016/j.cub.2009.02.056
In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy
Abstract
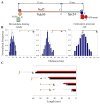
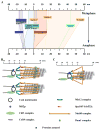
The kinetochore is a macromolecular protein machine [1] that links centromeric chromatin to the plus ends of one or more microtubules (MTs) and segregates chromosomes during cell division. Its core structure consists of eight multicomponent protein complexes, most of which are conserved in all eukaryotes. We use an in vivo two-color fluorescence microscopy technique to determine, for the first time, the location of these proteins along the budding yeast kinetochore axis at nanometer resolution. Together with kinetochore protein counts [2, 3], these localizations predict the 3D protein architecture of a metaphase kinetochore-microtubule attachment and provide new functional insights. We also find that the kinetochore becomes much shorter in anaphase as metaphase tension is lost. Shortening is due mainly to a decrease in the length of the Ndc80 complex, which may result either from intramolecular bending of the Ndc80 complex at the kink within the stalk region of the Ndc80-Nuf2 dimer [4, 5] or from a change in its orientation relative to the microtubule axis. Conformational changes within the Ndc80 and Mtw1 complexes may serve as mechanical cues for tension-dependent regulation of MT attachment and the spindle-assembly checkpoint. The geometry of the core structure of the budding yeast kinetochore reported here is remarkably similar to that found in mammalian kinetochores, indicating that kinetochore structure is conserved in eukaryotes with either point or regional centromeres.
Figures

Similar articles
-
The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity.Genes Dev. 2003 Jan 1;17(1):101-14. doi: 10.1101/gad.1040903. Genes Dev. 2003. PMID: 12514103 Free PMC article.
-
Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex.J Mol Biol. 2011 Jan 14;405(2):548-59. doi: 10.1016/j.jmb.2010.11.012. Epub 2010 Nov 12. J Mol Biol. 2011. PMID: 21075115 Free PMC article.
-
Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast.J Cell Biol. 2004 Feb 16;164(4):535-46. doi: 10.1083/jcb.200308100. Epub 2004 Feb 9. J Cell Biol. 2004. PMID: 14769859 Free PMC article.
-
Visualizing kinetochore architecture.Curr Opin Struct Biol. 2011 Oct;21(5):661-9. doi: 10.1016/j.sbi.2011.07.009. Epub 2011 Aug 22. Curr Opin Struct Biol. 2011. PMID: 21862320 Free PMC article. Review.
-
How the SAC gets the axe: Integrating kinetochore microtubule attachments with spindle assembly checkpoint signaling.Bioarchitecture. 2015;5(1-2):1-12. doi: 10.1080/19490992.2015.1090669. Epub 2015 Oct 2. Bioarchitecture. 2015. PMID: 26430805 Free PMC article. Review.
Cited by
-
Tension-dependent nucleosome remodeling at the pericentromere in yeast.Mol Biol Cell. 2012 Jul;23(13):2560-70. doi: 10.1091/mbc.E11-07-0651. Epub 2012 May 16. Mol Biol Cell. 2012. PMID: 22593210 Free PMC article.
-
Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore.Nat Rev Mol Cell Biol. 2013 Jan;14(1):25-37. doi: 10.1038/nrm3494. Nat Rev Mol Cell Biol. 2013. PMID: 23258294 Free PMC article. Review.
-
Unraveling the kinetochore nanostructure in Schizosaccharomyces pombe using multi-color SMLM imaging.J Cell Biol. 2023 Apr 3;222(4):e202209096. doi: 10.1083/jcb.202209096. Epub 2023 Jan 27. J Cell Biol. 2023. PMID: 36705602 Free PMC article.
-
The stoichiometry of the outer kinetochore is modulated by microtubule-proximal regulatory factors.J Cell Biol. 2019 Jul 1;218(7):2124-2135. doi: 10.1083/jcb.201810070. Epub 2019 May 22. J Cell Biol. 2019. PMID: 31118239 Free PMC article.
-
Visualization and identification of the structures formed during early stages of fibrin polymerization.Blood. 2011 Apr 28;117(17):4609-14. doi: 10.1182/blood-2010-07-297671. Epub 2011 Jan 19. Blood. 2011. PMID: 21248064 Free PMC article.
References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

