Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells
- PMID: 19345118
- PMCID: PMC3012962
- DOI: 10.1016/j.immuni.2009.01.014
Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells
Abstract
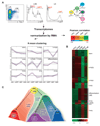
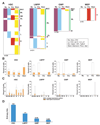
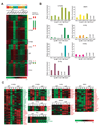
The mechanisms regulating lineage potential during early hematopoiesis were investigated. First, a cascade of lineage-affiliated gene expression signatures, primed in hematopoietic stem cells (HSCs) and differentially propagated in lineage-restricted progenitors, was identified. Lymphoid transcripts were primed as early as the HSC, together with myeloid and erythroid transcripts. Although this multilineage priming was resolved upon subsequent lineage restrictions, an unexpected cosegregation of lymphoid and myeloid gene expression and potential past a nominal myeloid restriction point was identified. Finally, we demonstrated that whereas the zinc finger DNA-binding factor Ikaros was required for induction of lymphoid lineage priming in the HSC, it was also necessary for repression of genetic programs compatible with self-renewal and multipotency downstream of the HSC. Taken together, our studies provide new insight into the priming and restriction of lineage potentials during early hematopoiesis and identify Ikaros as a key bivalent regulator of this process.
Figures

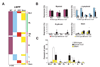

Comment in
-
A s-myly route toward lymphoid differentiation.Immunity. 2009 Apr 17;30(4):474-6. doi: 10.1016/j.immuni.2009.03.006. Immunity. 2009. PMID: 19371711
References
-
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. - PubMed
-
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. - PubMed
-
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. - PubMed
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

