Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner
- PMID: 19345191
- PMCID: PMC2673135
- DOI: 10.1016/j.cell.2009.01.040
Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner
Abstract
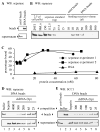
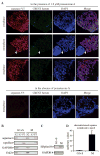
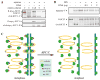
Sister chromatid separation is triggered by the separase-catalyzed cleavage of cohesin. This process is temporally controlled by cell-cycle-dependent factors, but its biochemical mechanism and spatial regulation remain poorly understood. We report that cohesin cleavage by human separase requires DNA in a sequence-nonspecific manner. Separase binds to DNA in vitro, but its proteolytic activity, measured by its autocleavage, is not stimulated by DNA. Instead, biochemical characterizations suggest that DNA mediates cohesin cleavage by bridging the interaction between separase and cohesin. In human cells, a fraction of separase localizes to the mitotic chromosome. The importance of the chromosomal DNA in cohesin cleavage is further demonstrated by the observation that the cleavage of the chromosome-associated cohesins is sensitive to nuclease treatment. Our observations explain why chromosome-associated cohesins are specifically cleaved by separase and the soluble cohesins are left intact in anaphase.
Figures

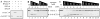


Similar articles
-
DNA-dependent cohesin cleavage by separase.Nucleus. 2010 Jan-Feb;1(1):4-7. doi: 10.4161/nucl.1.1.10010. Nucleus. 2010. PMID: 21327097 Free PMC article.
-
A single cohesin complex performs mitotic and meiotic functions in the protist tetrahymena.PLoS Genet. 2013 Mar;9(3):e1003418. doi: 10.1371/journal.pgen.1003418. Epub 2013 Mar 28. PLoS Genet. 2013. PMID: 23555314 Free PMC article.
-
The complete removal of cohesin from chromosome arms depends on separase.J Cell Sci. 2007 Dec 1;120(Pt 23):4188-96. doi: 10.1242/jcs.011528. Epub 2007 Nov 14. J Cell Sci. 2007. PMID: 18003702
-
What is your assay for sister-chromatid cohesion?EMBO J. 2007 Nov 14;26(22):4609-18. doi: 10.1038/sj.emboj.7601898. Epub 2007 Oct 25. EMBO J. 2007. PMID: 17962808 Free PMC article. Review. No abstract available.
-
The sister bonding of duplicated chromosomes.Semin Cell Dev Biol. 2011 Aug;22(6):566-71. doi: 10.1016/j.semcdb.2011.03.013. Epub 2011 Apr 7. Semin Cell Dev Biol. 2011. PMID: 21497666 Free PMC article. Review.
Cited by
-
Determinants of human cyclin B1 association with mitotic chromosomes.PLoS One. 2013;8(3):e59169. doi: 10.1371/journal.pone.0059169. Epub 2013 Mar 11. PLoS One. 2013. PMID: 23505570 Free PMC article.
-
Separase and Roads to Disengage Sister Chromatids during Anaphase.Int J Mol Sci. 2023 Feb 27;24(5):4604. doi: 10.3390/ijms24054604. Int J Mol Sci. 2023. PMID: 36902034 Free PMC article. Review.
-
Pan-Cancer analysis and experimental validation identify the oncogenic nature of ESPL1: Potential therapeutic target in colorectal cancer.Front Immunol. 2023 Mar 16;14:1138077. doi: 10.3389/fimmu.2023.1138077. eCollection 2023. Front Immunol. 2023. PMID: 37006282 Free PMC article.
-
MicroRNA-202 safeguards meiotic progression by preventing premature SEPARASE-mediated REC8 cleavage.EMBO Rep. 2022 Aug 3;23(8):e54298. doi: 10.15252/embr.202154298. Epub 2022 Jun 17. EMBO Rep. 2022. PMID: 35712867 Free PMC article.
-
Wapl is an essential regulator of chromatin structure and chromosome segregation.Nature. 2013 Sep 26;501(7468):564-8. doi: 10.1038/nature12471. Epub 2013 Aug 25. Nature. 2013. PMID: 23975099 Free PMC article.
References
-
- Akhmedov AT, Gross B, Jessberger R. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending. J Biol Chem. 1999;274:38216–38224. - PubMed
-
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. - PubMed
-
- Fan HY, Sun QY, Zou H. Regulation of Separase in Meiosis: Separase is Activated at the Metaphase I-II Transition in Xenopus Oocytes during Meiosis. Cell Cycle. 2006;5:198–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

