The postsynaptic density proteins Homer and Shank form a polymeric network structure
- PMID: 19345194
- PMCID: PMC2680917
- DOI: 10.1016/j.cell.2009.01.050
The postsynaptic density proteins Homer and Shank form a polymeric network structure
Abstract
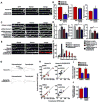
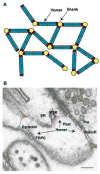
The postsynaptic density (PSD) is crucial for synaptic functions, but the molecular architecture retaining its structure and components remains elusive. Homer and Shank are among the most abundant scaffolding proteins in the PSD, working synergistically for maturation of dendritic spines. Here, we demonstrate that Homer and Shank, together, form a mesh-like matrix structure. Crystallographic analysis of this region revealed a pair of parallel dimeric coiled coils intercalated in a tail-to-tail fashion to form a tetramer, giving rise to the unique configuration of a pair of N-terminal EVH1 domains at each end of the coiled coil. In neurons, the tetramerization is required for structural integrity of the dendritic spines and recruitment of proteins to synapses. We propose that the Homer-Shank complex serves as a structural framework and as an assembly platform for other PSD proteins.
Figures

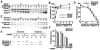



References
-
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. - PubMed
-
- Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143–154. - PubMed
-
- Boeckers TM, Liedtke T, Spilker C, Dresbach T, Bockmann J, Kreutz MR, Gundelfinger ED. C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J Neurochem. 2005;92:519–524. - PubMed
-
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. - PubMed
-
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

