APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein
- PMID: 19345973
- PMCID: PMC7103413
- DOI: 10.1016/j.virol.2009.03.010
APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein
Abstract
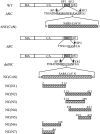
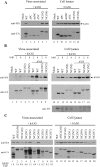
We previously reported that replacing HIV-1 nucleocapsid (NC) domain with SARS-CoV nucleocapsid (N) residues 2-213, 215-421, or 234-421 results in efficient virus-like particle (VLP) production at a level comparable to that of wild-type HIV-1. In this study we demonstrate that these chimeras are capable of packaging large amounts of human APOBEC3G (hA3G), and that an HIV-1 Gag chimera containing the carboxyl-terminal half of human coronavirus 229E (HCoV-229E) N as a substitute for NC is capable of directing VLP assembly and efficiently packaging hA3G. When co-expressed with SARS-CoV N and M (membrane) proteins, hA3G was efficiently incorporated into SARS-CoV VLPs. Data from GST pull-down assays suggest that the N sequence involved in N-hA3G interactions is located between residues 86 and 302. Like HIV-1 NC, the SARS-CoV or HCoV-229E N-associated with hA3G depends on the presence of RNA, with the first linker region essential for hA3G packaging into both HIV-1 and SARS-CoV VLPs. The results raise the possibility that hA3G is capable of associating with different species of viral structural proteins through a potentially common, RNA-mediated mechanism.
Figures



Similar articles
-
Severe acute respiratory syndrome coronavirus nucleocapsid protein confers ability to efficiently produce virus-like particles when substituted for the human immunodeficiency virus nucleocapsid domain.J Biomed Sci. 2008 Nov;15(6):719-29. doi: 10.1007/s11373-008-9265-8. Epub 2008 Jul 1. J Biomed Sci. 2008. PMID: 18592403 Free PMC article.
-
APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker.J Virol. 2007 May;81(10):5000-13. doi: 10.1128/JVI.02237-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344295 Free PMC article.
-
The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA.J Virol. 2007 Oct;81(20):11322-31. doi: 10.1128/JVI.00162-07. Epub 2007 Aug 1. J Virol. 2007. PMID: 17670826 Free PMC article.
-
Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors.Microbiol Mol Biol Rev. 2009 Jun;73(2):211-32. doi: 10.1128/MMBR.00040-08. Microbiol Mol Biol Rev. 2009. PMID: 19487726 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Self-assembly of severe acute respiratory syndrome coronavirus membrane protein.J Biol Chem. 2010 Apr 23;285(17):12862-72. doi: 10.1074/jbc.M109.030270. Epub 2010 Feb 12. J Biol Chem. 2010. PMID: 20154085 Free PMC article.
-
Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution.Virology. 2021 Apr;556:62-72. doi: 10.1016/j.virol.2020.12.018. Epub 2021 Jan 7. Virology. 2021. PMID: 33545556 Free PMC article. Review.
-
Viral N protein hijacks deaminase-containing RNA granules to enhance SARS-CoV-2 mutagenesis.EMBO J. 2024 Dec;43(24):6444-6468. doi: 10.1038/s44318-024-00314-y. Epub 2024 Nov 20. EMBO J. 2024. PMID: 39567830 Free PMC article.
-
SARS-CoV nucleocapsid protein interacts with cellular pyruvate kinase protein and inhibits its activity.Arch Virol. 2012 Apr;157(4):635-45. doi: 10.1007/s00705-011-1221-7. Epub 2012 Jan 6. Arch Virol. 2012. PMID: 22222284 Free PMC article.
-
A modular framework for the development of targeted Covid-19 blood transcript profiling panels.J Transl Med. 2020 Jul 31;18(1):291. doi: 10.1186/s12967-020-02456-z. J Transl Med. 2020. PMID: 32736569 Free PMC article.
References
-
- Alce T.M., Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279(33):34083–34086. - PubMed
-
- Bennett R.P., Presnyak V., Wedekind J.E., Smith H.C. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 2008;283(12):7320–7327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

